Viewing Gene Reports¶
Gene reports provide information related to the gene where selected variants reside. Available reports include:
Note
Available gene reports depend on the study type that was selected when the study was created (see also Creating a new study).
To access Gene reports, click on a gene symbol in any Variant listing view or in the header of the Variant Summary window, and select a gene report from the drop-down list.
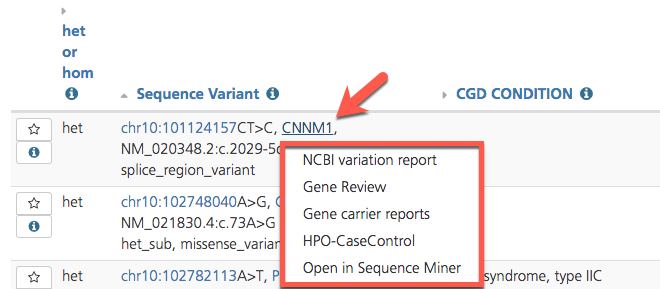
Accessing Gene reports from the Variant listing¶
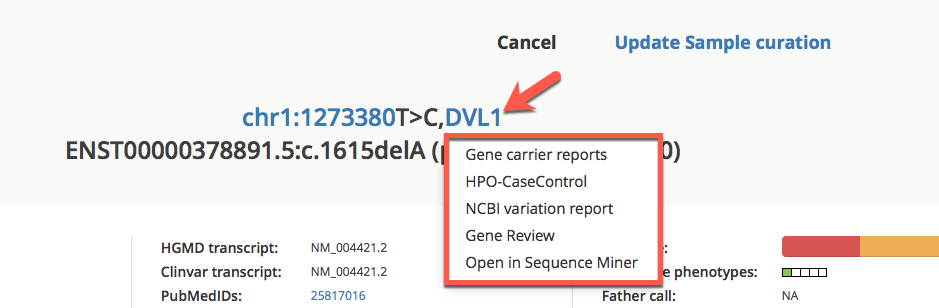
Accessing Gene reports from the Variant view¶
NCBI variation report¶
Selecting NCBI variation report opens the NCBI Variation Viewer in a new window, where you can view, search, and navigate known variants in the selected gene region. It displays a graphical representation of candidate gene transcripts. You can view the position(s) within the gene that contain known-SNV as well as sites of larger copy number variations. These variants contain annotations from ClinVar, dbSNP, and dbVAR.
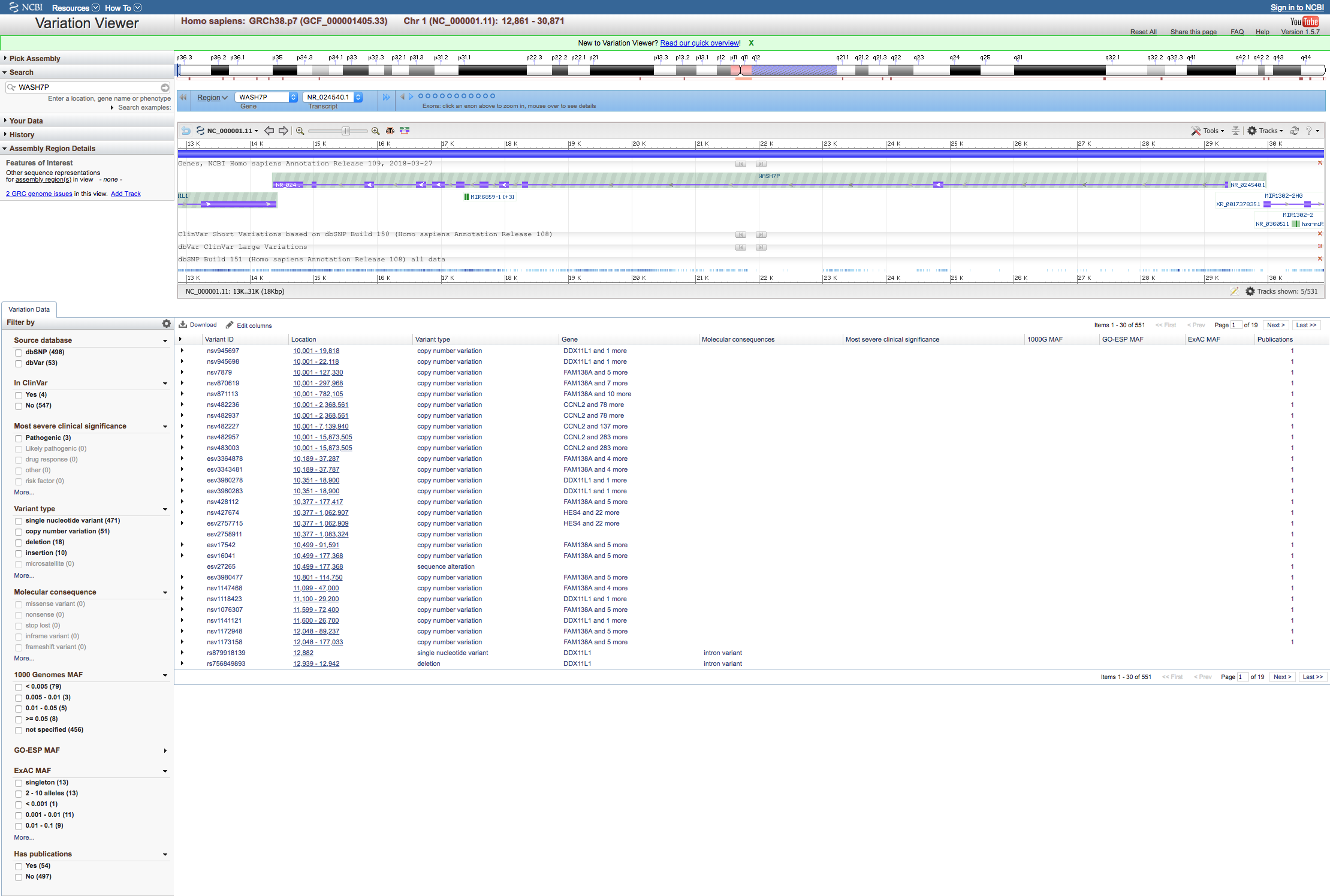
NCBI Variation Viewer¶
Gene review¶
Selecting Gene review opens an NCBI Bookshelf search for the selected gene in a new window, using the “[GeneSymbol]” search modifier.
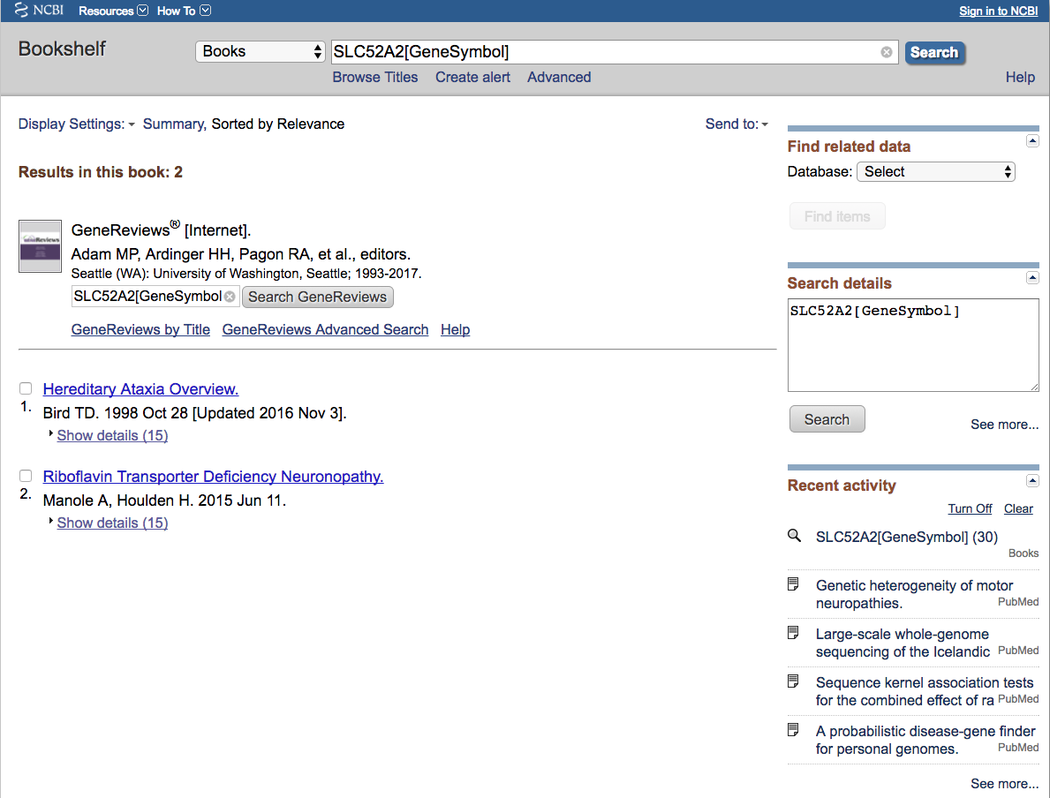
NCBI Bookshelf search¶
Gene carrier reports¶
Selecting Gene carrier reports opens a new window with available reports displayed in subtabs on the Carriers tab of the Gene context report view.
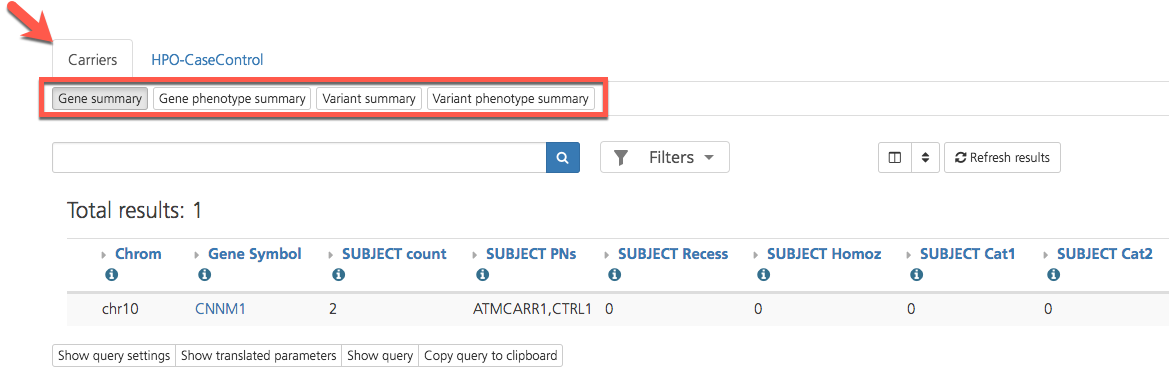
Gene carrier reports summarize individual carrier and phenotypic counts related to the selected gene and variants within the gene.
The following four carrier reports are available in CSA:
Gene summary - Summary of project participants that contain a variant in the gene
Gene phenotype summary - Summary of project participants with a registered phenotype that contain a variant in the gene
Variant summary - List of variants in the gene and number of project participants that contain each variant
Variant phenotype summary - List of variants in the gene and number of project participants with a registered phenotype
In general, each subtab provides a count of individuals identified as carrying a variant in the selected gene and information about how the identified variant has been annotated for each instance.
Results can be filtered by using the search bar and Filters menu at the top of the page.
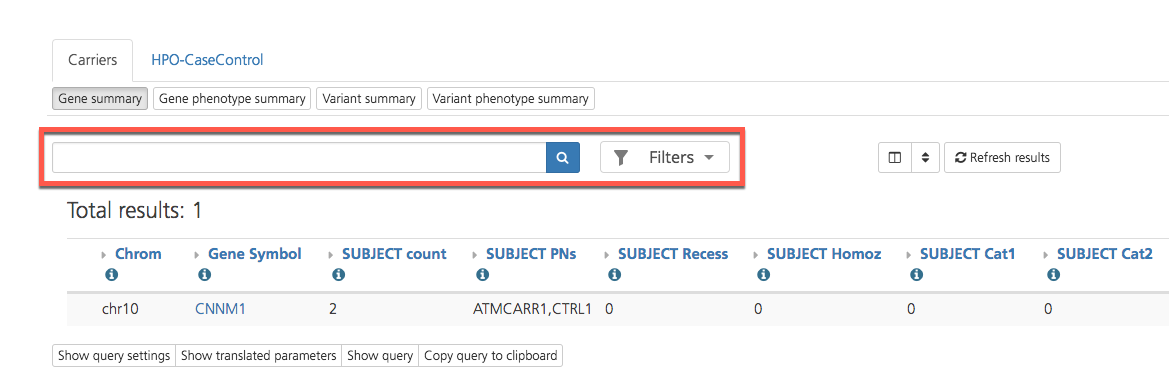
Gene summary¶
The Gene summary subtab displays the number of participants in the project that have the selected gene variant. Sample names are displayed along with the carrier status of the individuals in the project that have the indicated gene variant.

Gene summary report¶
Gene phenotype summary¶
The Gene phenotype summary subtab provides a count of the number of participants in the project that have a variant in the selected gene, as well as the associated phenotypic (HPO and/or OMIM) codes. Only participants with a registered phenotypic term are included in this table. Each row in the table is a set of phenotypic terms found in one or more participants.
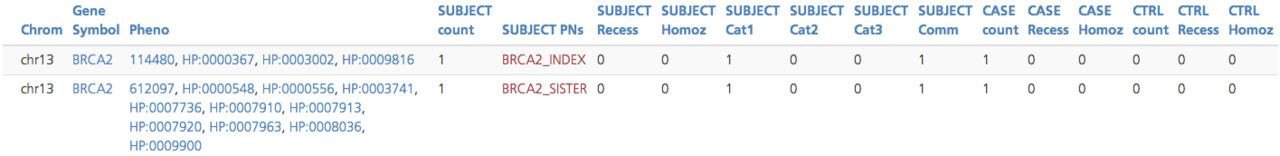
Gene phenotype summary report¶
Column descriptions¶
Gene summary and Gene phenotype summary report columns are described in the table below. You can also access column descriptions by clicking the Info icon next to each column header.
Column |
Description |
|---|---|
SUBJECT count |
The number of subjects in the database who have the variant in the gene (or this variant, depending on which report is selected) and the corresponding phenotype listed in the Pheno column |
SUBJECT PNs |
A list of all IDs for the subjects in the study who carry this variant and the corresponding phenotype listed in the Pheno column |
SUBJECT Recess |
The number of subjects in the database who are potentially recessive carriers for the gene, i.e., are homozgous for a variant or carry another variant in the gene (or this variant, depending on which report is selected), and have the corresponding phenotype listed in the Pheno column |
SUBJECT Homoz |
The number of subjects in the database who are homozygous carriers of the variant in the gene (or this variant, depending on which report is selected) and the corresponding phenotype listed in the Pheno column |
SUBJECT Cat1 |
The number of subjects in the database who have a Cat1 variant in the gene (or this variant, depending on which report is selected) and the corresponding phenotype listed in the Pheno column |
SUBJECT Cat2 |
The number of subjects in the database who have a Cat2 variant in the gene (or this variant, depending on which report is selected) and the corresponding phenotype listed in the Pheno column |
SUBJECT Cat3 |
The number of subjects in the database who have a Cat3 variant in the gene (or this variant, depending on which report is selected) and the corresponding phenotype listed in the Pheno column |
SUBJECT Comm |
The number of subjects in the database who have a variant in the gene (or this variant, depending on which report is selected) with user comments in the local knowledge base and the corresponding phenotype listed in the Pheno column |
CASE count |
The number of affected subjects in the study who have this variant and the corresponding phenotype listed in the Pheno column |
CASE Recess |
The number of affected subjects in the study who are potentially recessive carriers for the gene, i.e., are homozgous for this variant or carry another variant in the gene (or this variant, depending on which report is selected), and have the corresponding phenotype listed in the Pheno column |
CASE Homoz |
The number of affected subjects in the study who are homozygous carriers of this variant and the corresponding phenotype listed in the Pheno column |
CTRL count |
The number of unaffected subjects in the study who have this variant and the corresponding phenotype listed in the Pheno column |
CTRL Recess |
The number of unaffected subjects in the study who are potentially recessive carriers for the gene, i.e., are homozgous for this variant or carry another variant in the gene (or this variant, depending on which report is selected), and have the corresponding phenotype listed in the Pheno column |
CTRL Homoz |
The number of unaffected subjects in the study who are homozygous carriers of this variant and the corresponding phenotype listed in the Pheno column |
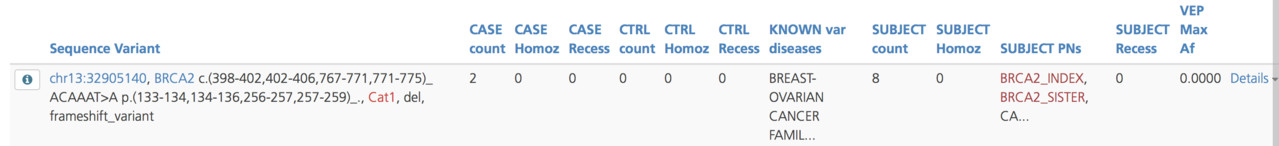
Variant summary¶
The Variant summary subtab lists the variants in the gene and displays a count of the participants that have each variant, known diseases associated with the selected gene variant, and the VEP maximum allele frequency reported for the selected gene variant.
To see information about the quality control of the variant call and phenotype (HPO and OMIM codes) for subjects identified in the project with the same variant, click the Details link on the right-hand side of the selected row.
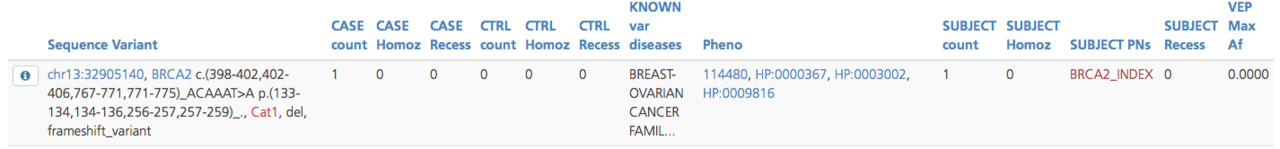
Variant phenotype summary¶
The Variant phenotype summary subtab lists the variants in the gene, along with a count of the number of participants with the variant and registered phenotypes in samples that contain the variant. Only participants with a registered phenotype term are included in this table. Variant listings are annotated with the participants containing the variant.
Clicking the Info icon next to the Sequence Variant opens the Variant view pop-up window, which displays information for All row attributes, Frequencies, and Transcripts on separate tabs (see also Variant annotation).
Column descriptions¶
Variant summary and Variant phenotype summary report columns are described in the table below. You can also access column descriptions by clicking the Info icon next to each column header.
Column |
Description |
|---|---|
CASE count |
The number of affected subjects in the study who have this variant and the corresponding phenotype listed in the Pheno column |
CASE Homoz |
The number of affected subjects in the study who are homozygous carriers of this variant and the corresponding phenotype listed in the Pheno column |
CASE Recess |
The number of affected subjects in the study who are potentially recessive carriers for the gene, i.e., are homozgous for this variant or carry another variant in the gene (or this variant, depending on which report is selected), and have the corresponding phenotype listed in the Pheno column |
CTRL count |
The number of unaffected subjects in the study who have this variant and the corresponding phenotype listed in the Pheno column |
CTRL Homoz |
The number of unaffected subjects in the study who are homozygous carriers of this variant and the corresponding phenotype listed in the Pheno column |
CTRL Recess |
The number of unaffected subjects in the study who are potentially recessive carriers for the gene, i.e., are homozgous for this variant or carry another variant in the gene (or this variant, depending on which report is selected), and have the corresponding phenotype listed in the Pheno column |
KNOWN var diseases |
Diseases known to be associated with the variant |
SUBJECT count |
The number of subjects in the database who have the variant in the gene (or this variant, depending on which report is selected) and the corresponding phenotype listed in the Pheno column |
SUBJECT Homoz |
The number of subjects in the database who are homozygous carriers of this variant and the corresponding phenotype listed in the Pheno column |
SUBJECT PNs |
A list of all IDs for the subjects in the study who carry this variant and the corresponding phenotype listed in the Pheno column |
SUBJECT Recess |
The number of subjects in the database who are potentially recessive carriers for the gene, i.e., are homozgous for a variant or carry another variant in the gene (or this variant, depending on which report is selected), and have the corresponding phenotype listed in the Pheno column |
VEP Max Af |
Maximum reported allele frequency from multiple sources available (e.g., 1000 Genome Project, Exome Variant server, ExAC, Iceland, Kyoto, etc., see also popAlleleFreq) |
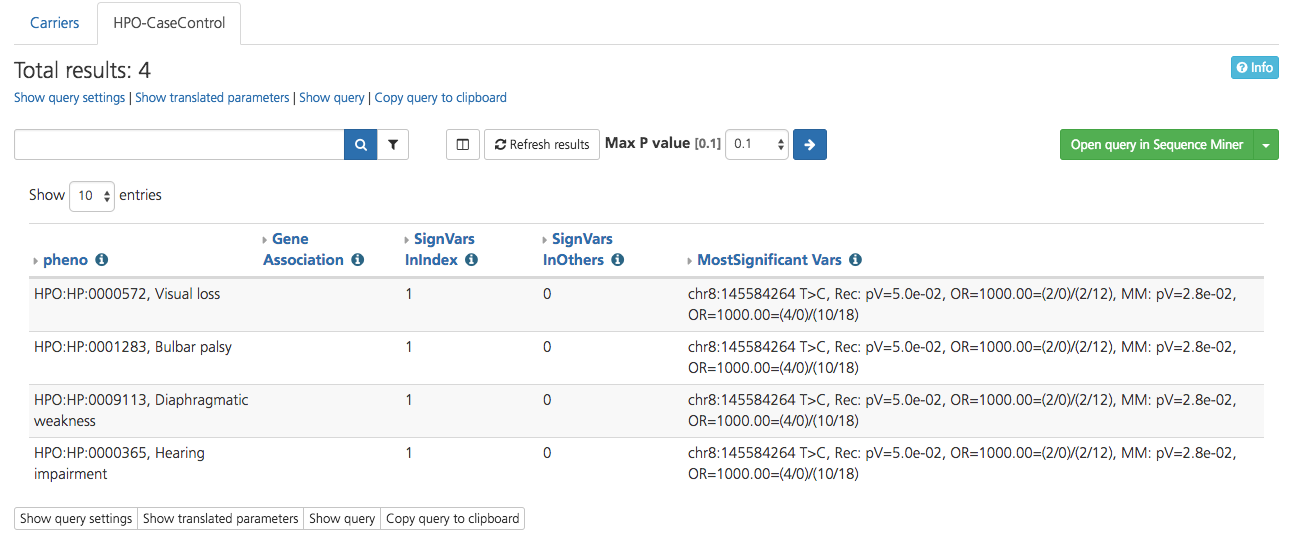
HPO-CaseControl¶
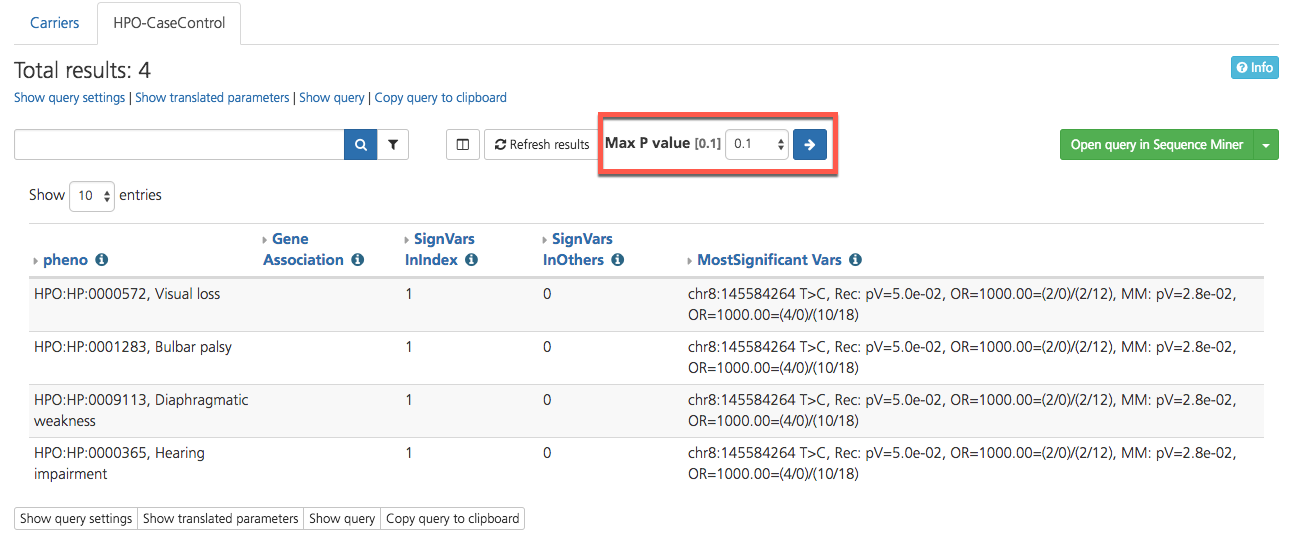
Selecting HPO-CaseControl opens opens a new window with results displayed on the HPO-CaseControl tab of the Gene context report view.
The HPO-CaseControl report runs a Case-Control analysis for all single codes phenotypes available in CSA. The output of the analysis determines the most significant variants determined by phenotype.
The report calculates an odds ratio (OR) and a p-value, taking into account the number of cases and controls that harbor a given phenotype. An OR and p-value is calculated for the predicted model of inheritance (Dominant, Recessive) and for the Multiplicative model.
Significant HPO terms are reported in the pheno column, along with genes associated with the phenotype (Gene Association), and the number of significant variants in the designated index case for the study (SignVarsinIndex) compared to the number of significant variants in other members of the study (SignVarsInOthers). The most significant variants identified are reported in the MostSignificantVars column.
Results can be filtered by using the search bar and Filters menu at the top of the page. Adjust the p-value by selecting a value from the drop-down list and clicking the arrow.
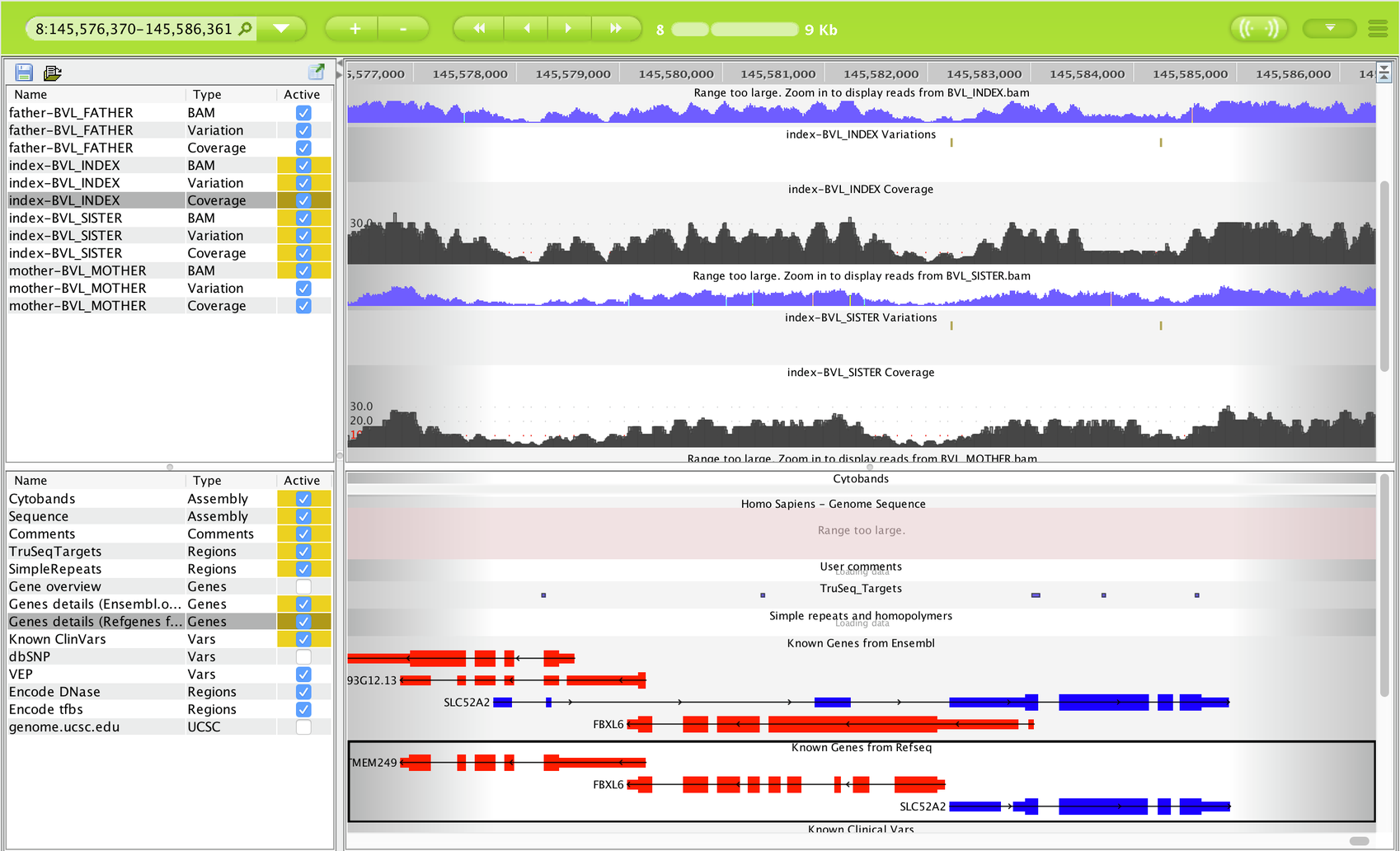
Open in Sequence Miner¶
Selecting Open in Sequence Miner opens the Genome Browser, which provides a visualization of the aligned read coverage and variants call for the selected gene across the individuals. Annotation tracks are also visible for the range of the gene, and include options to view regions of conservation through the UCSC genome browser window (see also Opening Sequence Miner).