Variant Curation¶
Adding a new variant curation¶
A new variant curation can be created from either the Variant Information or the Variant Curation window.
Initiating a new variant curation¶
From the Variant Information window¶
The Variants page is shown below. To open the Variant Information window for a selected variant by clicking the icon indicated below next to the variant in the Variant Listing.
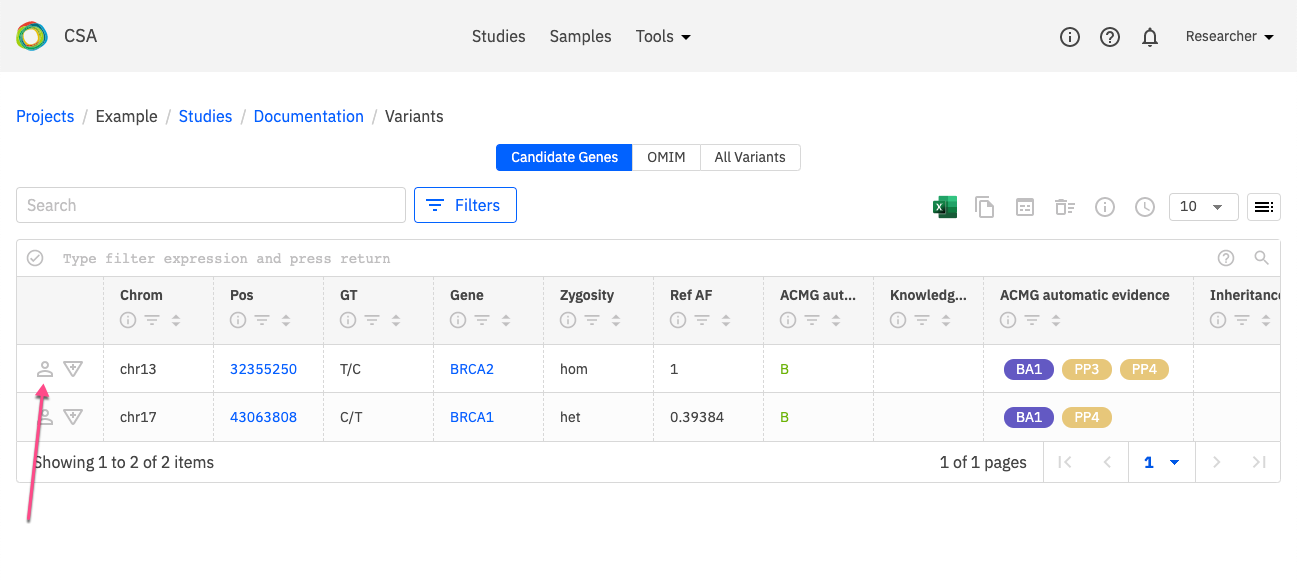
The Variant Information window will open as shown below. Click on Go to variant scoring to open the Variant Curation window.
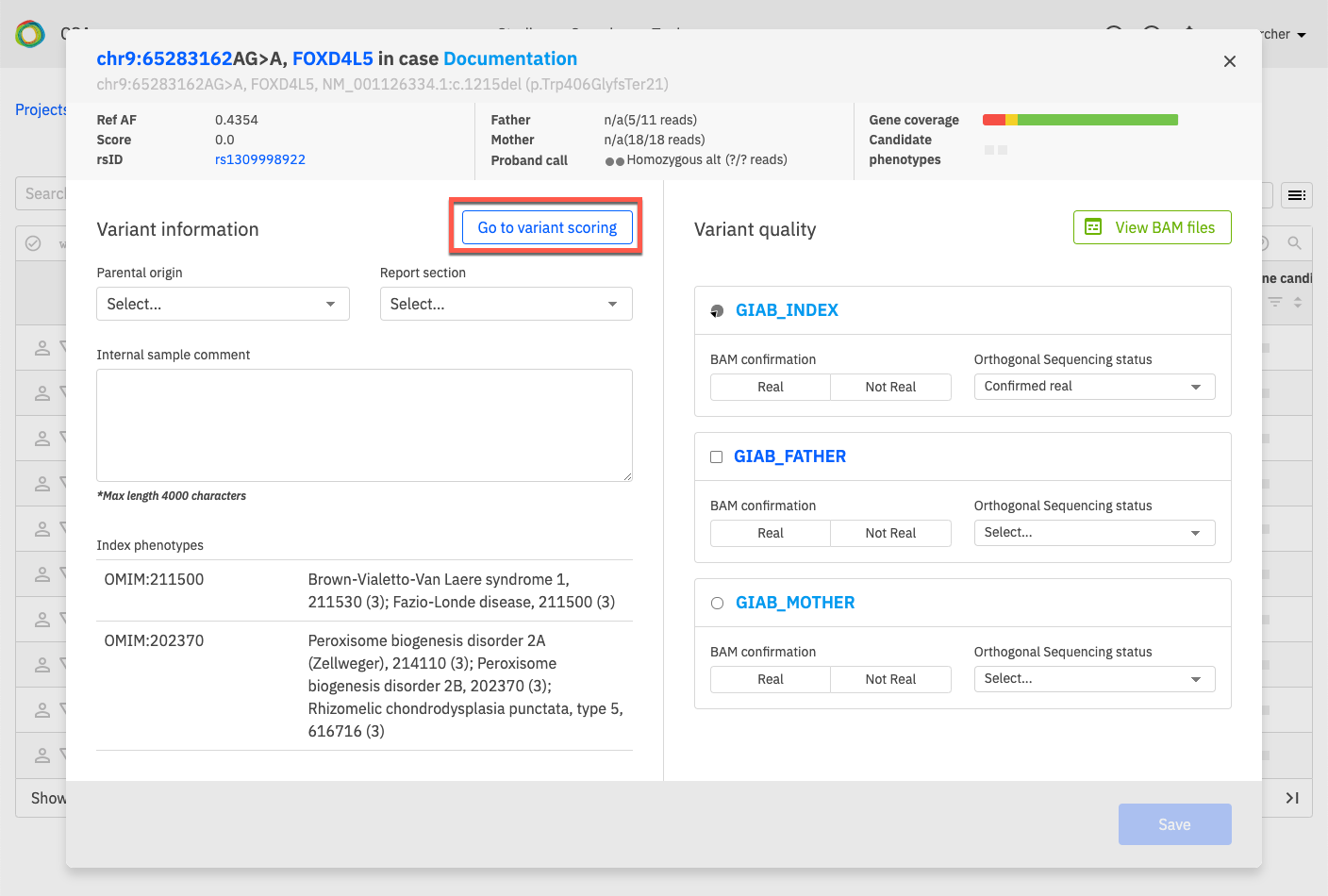
Finally, open the Add variant disease curation drop-down menu and select New variant disease curation.

This opens the New curation window for the selected variant.
From the Variant Curation window¶
The variant curation process can be started more directly through the Variants table itself. In the Variants page, click on the icton indicated below to open the Variant Curation window.
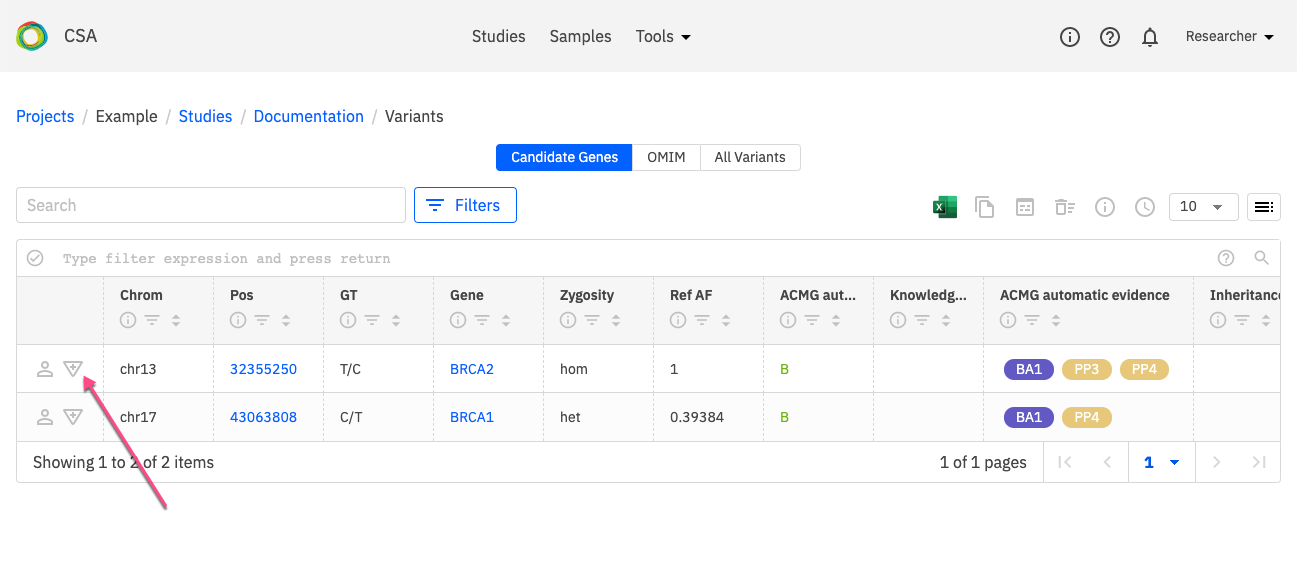
Open the Existing variant disease curations drop-down menu and select New variant disease curation.
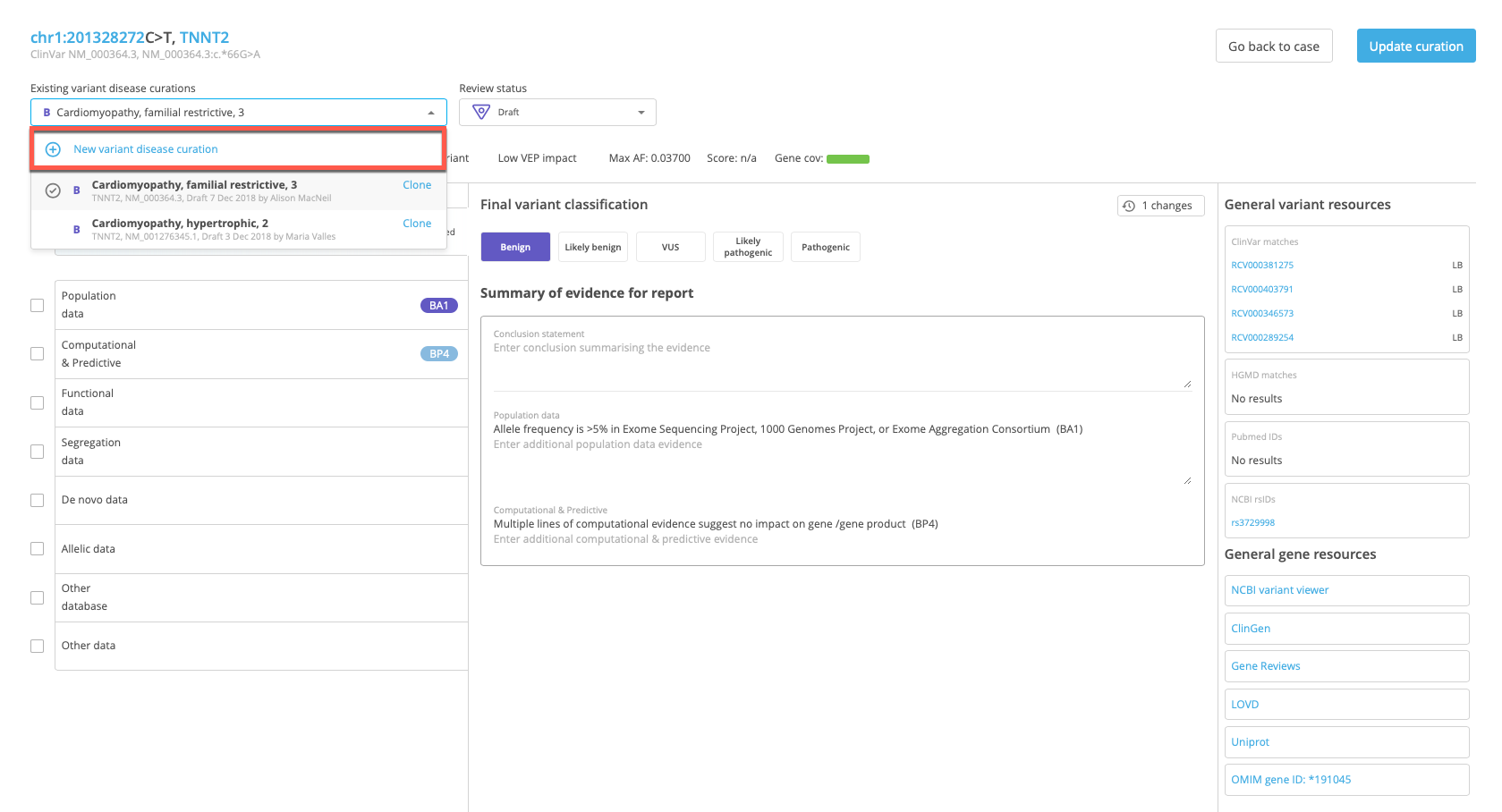
This opens the New curation window for the selected variant.
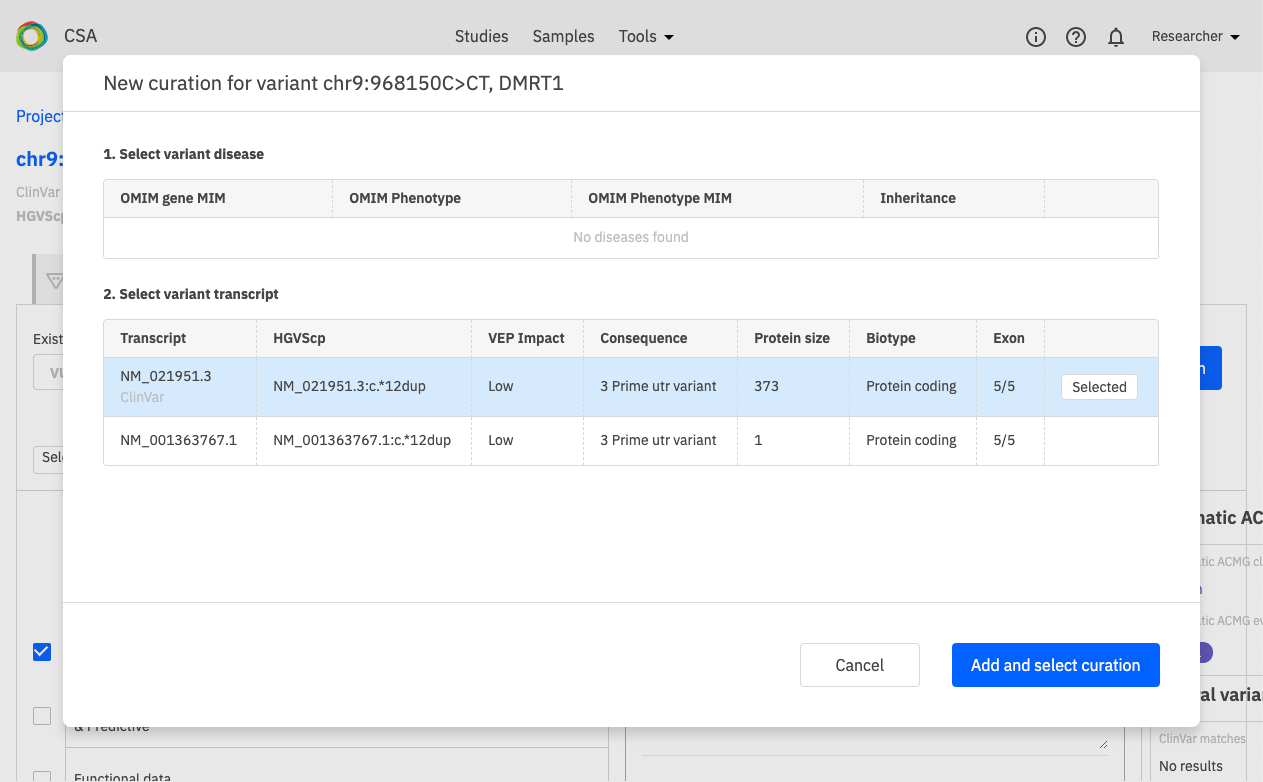
Defining a new variant creation¶
In the New curation window, define the new variant curation by selecting the disease and transcript.
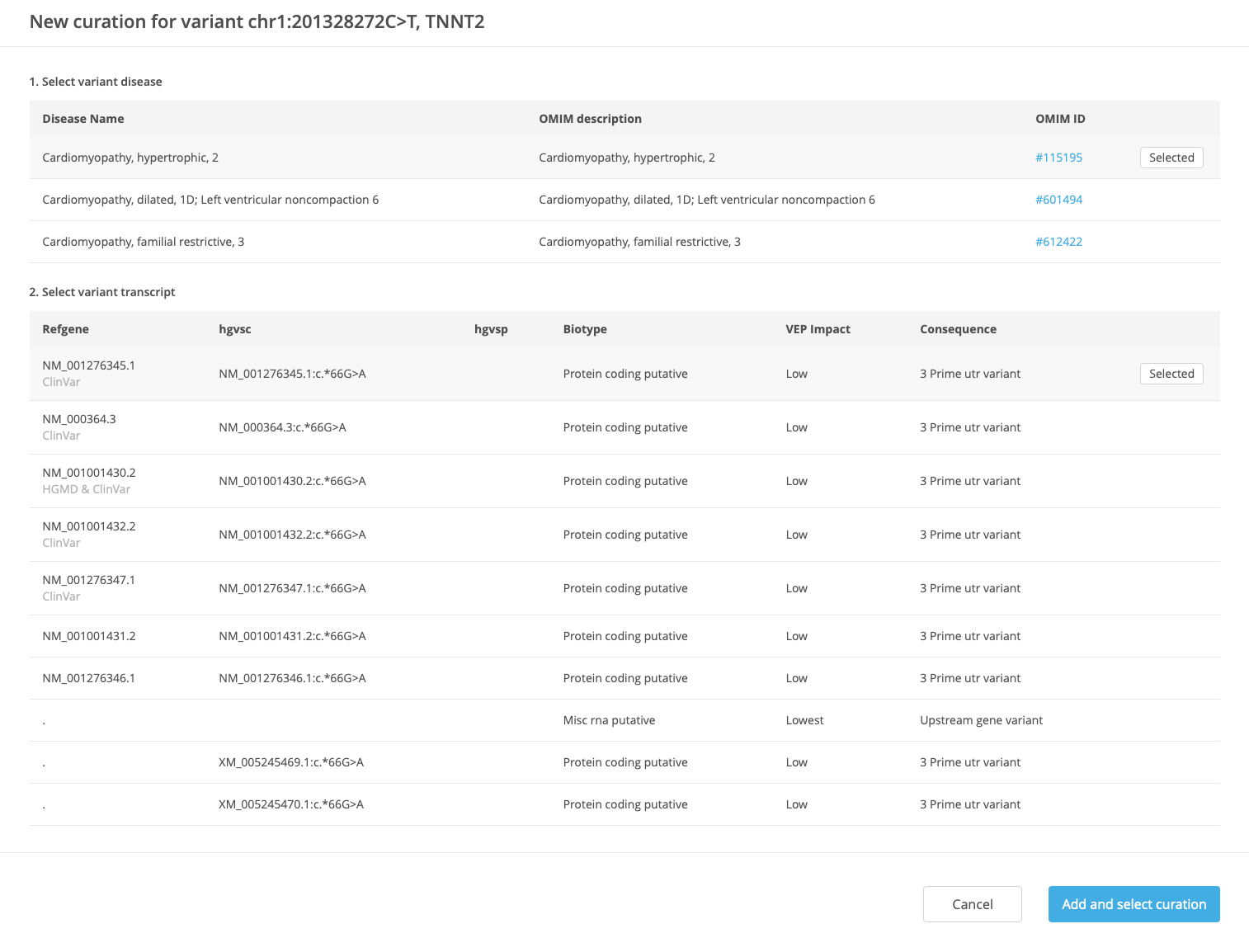
Each variant curation is uniquely defined by VDT:
Variant identity (position, ref string, alternate string, gene, and genome build)
Disease
Transcript
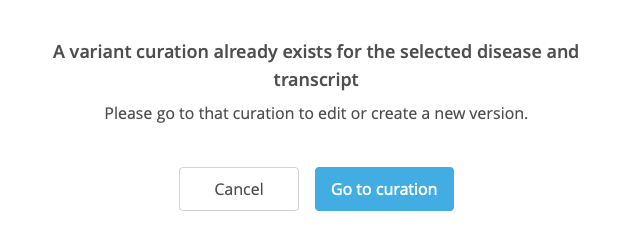
If the selected combination of VDT already exists, a prompt is displayed to redirect to the existing curation.
Diseases available for selection are based on OMIM disease entries associated with the current gene. The default disease for selection may be defined by a mapping file that indicates a preferred disease for each gene. If the current gene does not have any OMIM disease entries associated with it, the disease selection is blank (no disease available).
Transcripts available for selection are based on output from the VEP algorithm. The default transcript for selection is defined by HGMD and ClinVar preferred transcripts.
Once the disease and transcript are selected, click Add and select curation to save the curation and open the Variant Curation window (see also Editing a Variant Curation).
Editing a Variant Curation¶
Existing variant curations can be edited from the Variant Curation window, which is opened from either the Variant Summary window or upon creating a new variant curation.
The Variant Curation window consists of a Variant curation header that contains basic information about the selected variant, and tools for Variant curation scoring.
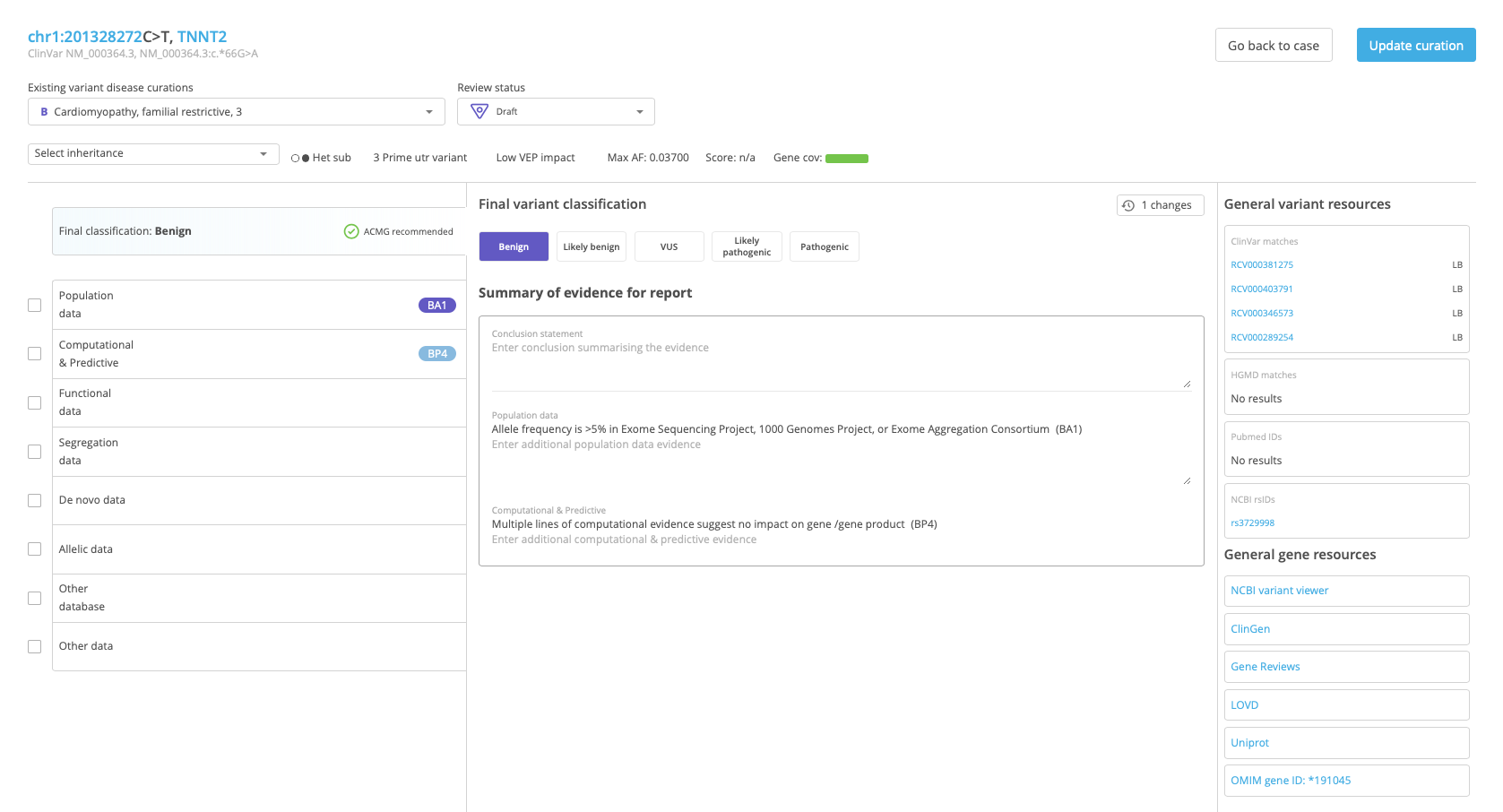
Variant curation header¶
The Variant curation header includes the following fields:
Variant identity
Existing variant disease curations
Review status
Inheritance
Variant details

Variant identity¶
This field displays the variant chromosome, position, reference nucleotide string, alternate nucleotide string, and the gene in which the variant is found.
Clicking the variant chromosome:position or gene symbol opens a drop-down menu. For example, selecting Open in Sequence Miner in the drop-down opens the Sequence Miner Genome Browser to show the aligned reads for all participants of the current study.
This section also displays an HGVS string, including the transcript ID (which identifies the transcript selected for this variant curation), change in the transcript, and resulting change in the protein. This HGVS format is derived from VEP resources.
Existing variant disease curations¶
This field displays the disease for the current variant curation.
Open the drop-down menu to select from existing variant curations for the selected variant or to create a new variant curation.
Each variant curation is uniquely defined by VDT:
Variant identity (position, ref string, alternate string, gene, and genome build)
Disease
Transcript
Selecting New variant disease curation opens a New curation window, where disease and transcript can be selected and added to the curation. If the selected combination of VDT already exists, a prompt is displayed to redirect to the existing curation (see also Adding a new variant curation).
Selecting Clone next to an existing variant disease curation copies the annotations from the existing variant curation to a new variant curation.
Review status¶
This field displays the current review status of the variant curation.
Open the drop-down menu to select a new review status. The status “Accepted” is available only to users with the Variant Approver user role. If the Review status is set to “Accepted” or “Expired”, the variant curation is locked from further editing. A new draft must be created to apply further edits.
The most typical workflow for variant curation review is Pending → Draft → Accepted. Available Review status options are described in the following table:
Icon |
Review status |
Description |
|---|---|---|
Accepted |
The variant curation has been reviewed and approved by a user with the Variant Approver user role. For users without the Variant Approver user role, this option is inactive. Once a variant curation is set to “Accepted”, the curation is locked from editing unless a new draft is created. |
|
Pending |
The variant curation is actively being worked on. This status is functionally the same as “Draft”, but indicates that the curation is still being worked on. |
|
Draft |
The variant curation is saved in a draft state. This status is functionally the same as “Pending”, but indicates the curation is not currently being worked on. |
|
Ignored |
The variant curation is labeled to avoid further attention. |
|
Expired |
The variant curation is due for another round of review. This status can be set manually. If a variant curation is set to “Expired”, the curation is locked from editing unless a new draft is created. |
Inheritance¶
Open the drop-down menu to select the inheritance for the current variant curation. The default inheritance value is derived from the OMIM entry for the selected disease. This value is stored as part of the current variant curation entry.
Variant details¶
This section displays various details of the variant in the context of the current case, including variant type (zygosity and substitution/insertion/deletion), VEP consequence and impact, Max AF, Max score (derived from Computational and predictive scores from the dbNSFP resource), and Gene coverage (average coverage across the current gene).
Variant curation scoring¶
The lower panels of the Variant Curation window are focused on variant curation scoring. These panels include:
Final classification
ACMG categories of evidence
Lines of evidence
Resources
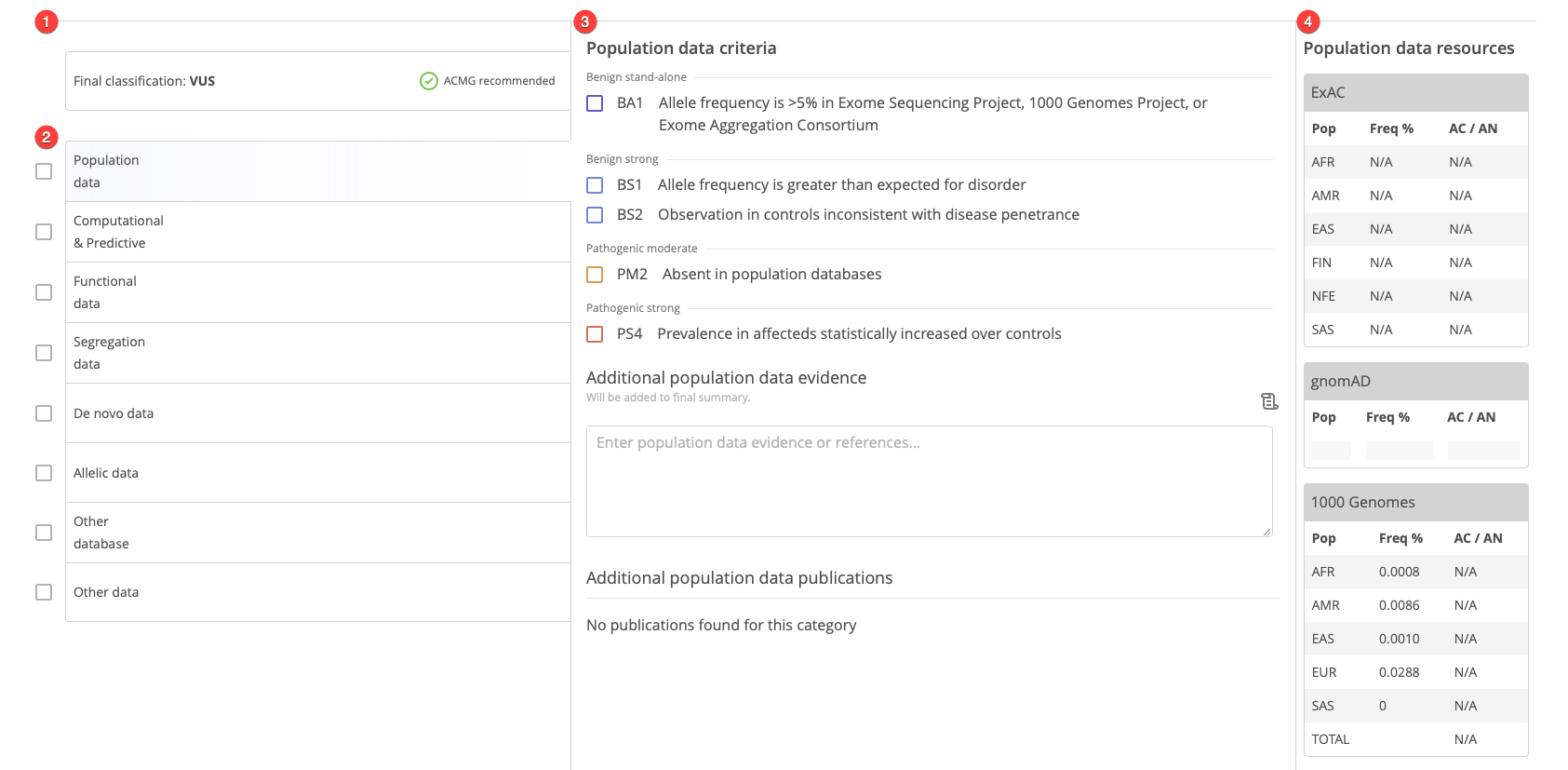
Final classification¶
The Final classification field displays the final classification for the variant curation. Clicking on this field reveals additional information and options.
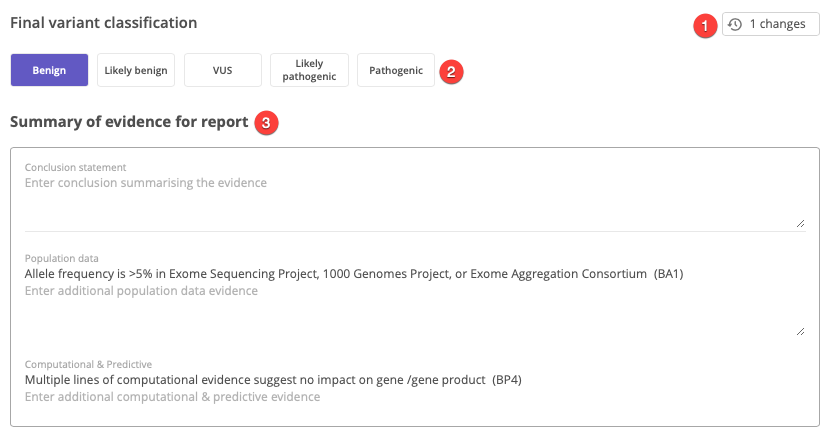
The final classification can be compared to the ACMG recommended classification, which is calculated according to the ACMG scoring guidelines based on the selected lines of evidence. The final classification may differ from the ACMG recommended classification if a custom score or a custom classification are selected.
Clicking inside the Final classification field activates the Final variant classification panel in the center panel, which includes the following:
Change log - Displays a record of changes for the variant curation
Custom classification - Provides an option to select a custom classification. Selecting a custom classification triggers a warning message the the ACMG recommended classification has be overridden.
Summary of evidence for the report - Includes a text box for entering a conclusion statement, as well as free-text entry fields for each category of evidence.
ACMG categories of evidence¶
Each vertical tab displays one of the categories of evidence defined by the ACMG scoring guidelines. Selecting an ACMG category of evidence displays the corresponding lines of evidence (center panel) and resources (rightmost panel).
The checkboxes next to each category of evidence can be used to track progress through the scoring process.
Lines of evidence¶
For each category of evidence, the following options can be selected in the center panel:
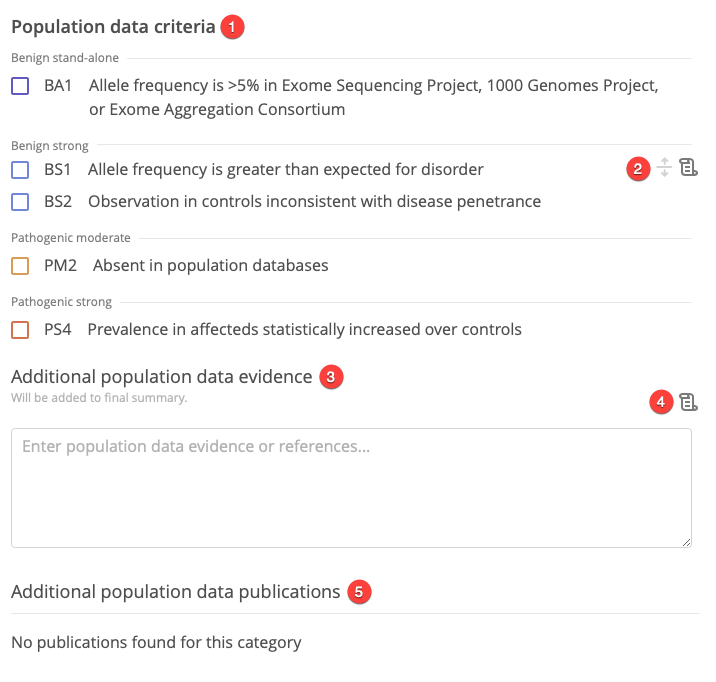
Select available lines of evidence
For each category, select lines of evidence by selecting the corresponding checkboxes. Selecting a line of evidence updates the category labels in the leftmost panel and the final classification score.
Apply custom weights
If appropriate, custom weights can be applied to each line of evidence. Selecting a custom weight for any line of evidence prompts a warning message in the Final variant classification panel.
Add summary information
For each category of evidence, notes can be added in the Evidence or references text box. Text entered here will also appear in the Final variant classification panel.
Attach publications
Attach publications for individual lines of evidence or the whole category of evidence by clicking the Publications icon.
In the Attach publication dialog, enter keywords into the search box to return text-matched, hyperlinked results from PubMed.
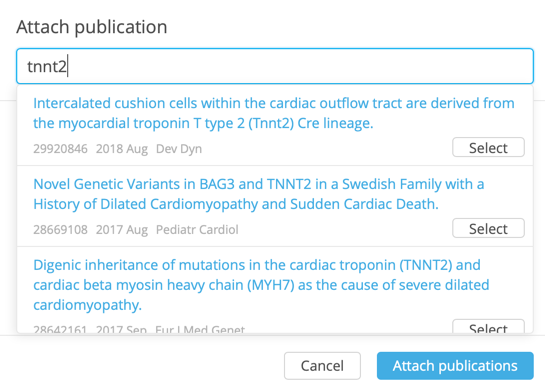
Click Select to link to the corresponding entry in PubMed.
To add the publication(s) to the current category of evidence, click Attach publications.
Attached publications can be managed from the Additional publications section in the center panel. Remove attached publications by deselecting all lines of evidence and categories.
Resources¶
For each category of evidence, the Resources panel on the right-hand side displays relevant resources collected for ease of scoring:
Category |
Resources |
|---|---|
Population data |
Include tables of allele frequencies and counts for population databases including ExAC, gnomAD, and 1000 Genomes |
Computational and predictive |
ClinVar and HGMD database matches, as well as Polyphen-2, SIFT, and DeepCODE |
Functional data |
ClinVar matches and OMIM database matches PubMed IDs |
Segregational data |
Gene carrier report for a summary of variant carrier status across all individuals in the project |
De novo data |
Zygosity, alternate allele depth, and coverage information for parents, if available |
Allelic data |
A link to view the aligned reads, and a summary of compound het information |
Other database |
ClinVar and HGMD database matches |
Other resources |
Gene carrier report for a summary of variant carrier status across all individuals in the project |