Reference Data¶
The ref folder (accessible from the File Explorer in Sequence Miner contains a collection of reference files, curated from a variety of database and combined into GOR format.
GORdb stores data indexed by genomic locus coordinates, thereby facilitating real-time retrieval and integration of clinical sequence data with annotation from public, proprietary and custom resources. The raw sequence data and annotation data are stored separately so changes to (e.g., addition of new sample data to the database or the release of a new reference package) can be handled without the need to rewrite per-sample annotation files. As reference data is updated, data joins are performed on the fly using the most up-to-date reference data.
A full list of reference data sources is shown in the table at the end of this page.
Each Clinical Sequence Analyzer (CSA) instance is deployed with its own build of the reference data and each CSA project gets a build setting when it is created. Studies within each project can reference different major-minor versions of the Reference Data build.
The reference data that is deployed with your instance of CSA may depend on your subscriptions to various banks of genomic data.
Reference data files¶
The following table provides a brief overview of selected reference files and directories:
Reference file |
Description |
|---|---|
ref/cancer/* |
Directory contains cancer-related files with clinical actionable information, commercial cancer panels, TCGA cancer related genes, CGD genes |
ref/dbsnp/* |
Directory contains variants from the dbSNP database |
ref/deepCODE/* |
Directory contains variants with scores from the deepCODE algorithm |
ref/disgenes/* |
Directory contains disease-related gene map files, including the ACMG minimum panel, CGD panel, immuno-related disease panel, the Kingsmore childhood panel, etc. |
ref/disvariants/* |
Directory contains variants from Clinvar and HGMD |
ref/encode/* |
Directory contains positions with associated ENCODE data |
ref/ensgenes/* |
Directory contains Ensembl gene-related infomation, exons, transcripts, pathway, Gene Ontology (GO), paralogs, etc. |
ref/hgmd/* |
Directory contains HGMD-related files, including variant details with clinical information and URL links |
ref/refgenes/* |
Directory contains the RefSeq gene related information, exons, transcripts, etc. |
ref/regulation/* |
Directory contains regulation-related files from ENCODE, etc. |
ref/repeats/* |
Directory contains files that identify regions of simple repeats |
ref/variants/* |
Directory contains variant information including data from population studies (EVS, ExAC, 1000 Genomes, etc.) |
ref/cancer_variants.gorz |
Variants from COSMIC and NCI-60 database |
ref/clinical_genes.gorz |
Disease genes based on variants from HGMD, ClinVar, and OMIM |
ref/clinical_variants.gorz |
Clinical variants from HGMD, ClinVar, and OMIM |
ref/genes.gorz |
Ensembl gene list with one entry per gene symbol |
ref/rgenes.gorz |
RefSeq gene list |
ref/1000G.gorz |
Variants with allele frequencies from the 1000 Genomes |
ref/dbnsfp.gorz |
Variants and annotations from the dbNSFP database |
ref/evs_anno.gorz + ref/evs_freq |
Variants with allele frequencies from the EVS database |
ref/exac.gorz |
Variants with allele frequencies from the EXAC database |
ref/freq_max.gorz |
Combined variants from EVS, 1000 Genomes, the Japanese Ancestry population from the Kyoto Consortium, the deCODE population survey of Iceland, Genomes of the Netherlands (GoNL), and ExAC |
ref/jpt_freq.gorz |
Variants with allele frequencies from the Japanese Ancestry population from the Kyoto Consortium |
ref/version.txt |
The version of databases listed in the ref folder |
Reference data sources¶
The following table contains a comprehensive list of all sources of the reference data. To find out the exact versions included in your installation of CSA, please refer to the version.txt file in the ref folder or to the release notes for your installation.
Resource |
Description |
|---|---|
RefSeq |
The RefSeq collection provides a comprehensive, integrated, non-redundant, well-annotated set of sequences, including genomic DNA, transcripts, and proteins. |
Ensembl |
The Ensembl project produces genome databases for vertebrates and other eukaryotic species, and makes this information freely available online. |
dbSNP |
Providing variants with accession numbers in the form of RS IDs, the NCBI dbSNP database integrates most germline variants. The allele frequency information in the database is provided directly from 1000 genomes, but dbSNP contains variants from many other sources as well. |
1000 Genomes |
The 1000 Genomes Project has sequenced over 1000 individuals from 14 populations by combining whole genome sequencing and whole exon sequencing. |
dbNSFP |
Non-Synonymous Function Predictions database annotates SNPs in the human genome with the functional predictions. |
EVS/ESP |
The Exome Variant Server (EVS) contains exome sequencing variants as part of the NHLBI Exome Sequencing Project (ESP).The ESP6500 has collected over 6500 exomes, including health controls, specific diseases. The goal of the ESP dataset is to release the frequency counts of specific variants without regard to phenotype. |
EXAC |
The Exome Aggregation Consortium collected data from unrelated individual exomes sequenced as part of various disease-specific and population genetic studies. |
COSMIC |
Catalogue of Somatic Mutation in Cancer project stores somatic mutation information and related details and contains information relating to human cancer. |
TCGA |
The Cancer Genome Atlas database provides somatic mutations and related disease information for a list of specific cancers. |
HGMD |
The Human Gene Mutation Database represents an attempt to collate known (published) gene lesions responsible for human inherited disease. |
CGD |
Clinical Genomic Database is a manually curated database of conditions with known genetic causes, focusing on medically significant genetic data with available interventions. |
OMIM |
Online Mendelian Inheritance in Man is a continuously updated catalog of human genes and genetic disorders and traits, with particular focus on the molecular relationship between genetic variation and phenotypic expression. |
ClinVar |
ClinVar is a freely accessible, public archive of reports of the relationships among human variations and phenotypes, with supporting evidence. |
ACMG |
American College of Medical Genetics and Genomics has recommended sets of genes for reporting incidental findings in clinical exome and genome sequencing. |
Population allele frequencies¶
There are several population surveys included in Clinical Sequence Analyzer (CSA). The following table summarizes the number of samples used to generate the allele frequencies for each database.
Database |
Number of exomes/genomes |
|---|---|
1000 Genomes |
2,577 |
EVS (ESP) |
6,503 |
ExAC |
60,706 |
gnomAD |
138,632 |
Genome of the Netherlands (GoNL) |
769 |
Kyoto Japanese |
1,208 |
Icelandic |
2,636 |
SUM |
212,958 |
Population survey data for a selected variant is also displayed in the References panel of the Variant Curation in CSA when the Population data category of evidence is selected for scoring.
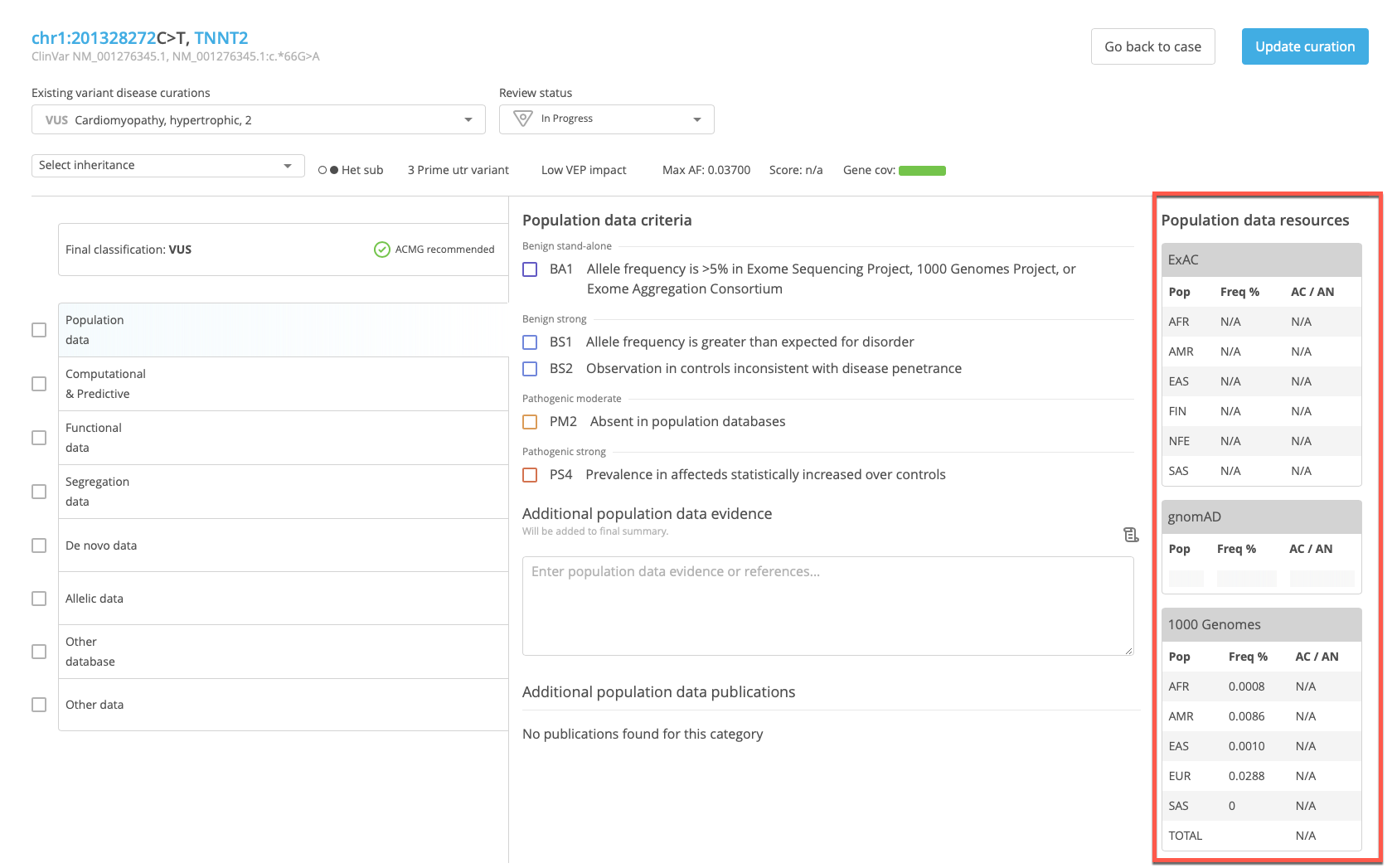
The tables in the population data references panel include the following information:
Pop - Population
Freq - Allele frequency
AC/AN - Allele count/allele number (the number of times the variant allele appears in a given population/the total number of times the allele is present in either the variant or reference sequence)
Following are brief descriptions of each available database.
ExAC¶
The ExAC table provides information from the following populations:
AFR - African/African American
AMR - Latino
EAS - East Asian
FIN - Finnish
NFE - Non-Finnish European
SAS - South Asian
Information provided by ExAC (Exome Aggregation Consortium) about population survey sizes is shown in the following table:
Population |
Male samples |
Female samples |
Total |
|---|---|---|---|
African/African American (AFR) |
1,888 |
3,315 |
5,203 |
Latino (AMR) |
2,254 |
3,535 |
5,789 |
East Asian (EAS) |
2,016 |
2,311 |
4,327 |
Finnish (FIN) |
2,084 |
1,223 |
3,307 |
Non-Finnish European (NFE) |
18,740 |
14,630 |
33,370 |
South Asian |
6,387 |
1,869 |
8,256 |
Other (OTH) |
275 |
179 |
454 |
TOTAL |
33,644 |
27,062 |
60,706 |
For more information, visit the ExAC Browser and ExAC FAQ.
gnomAD¶
The gnomeAD table provides information from the following populations:
AFR - African/African American
AMR - Latino
ASJ - Ashkenazi Jewish
EAS - East Asian
FIN - Finnish
NFE - Non-Finnish European
SAS - South Asian
TOTAL - Total variant allele frequency in the combined populations
The gnomAD database (Genome Aggregation Database) includes 123,136 exome samples and 15,496 whole genome samples. Information provided by gnomAD about the populations included in the database is shown in the following table:
Population |
Exomes |
Genomes |
Total |
|---|---|---|---|
African/African American |
7,652 |
4,368 |
12,020 |
Latino (AMR) |
16,791 |
419 |
17,210 |
Ashkenazi Jewish (ASJ) |
4,925 |
151 |
5,076 |
East Asian |
8,624 |
811 |
9,435 |
Finnish (FIN) |
11,150 |
1,747 |
12,897 |
Non-Finnish European (NFE) |
55,860 |
7,509 |
63,369 |
South Asian (SAS) |
15,391 |
0 |
15,391 |
Other (OTH) |
2,743 |
491 |
3,234 |
TOTAL |
123,136 |
15,496 |
138,632 |
The first release of gnomAD was known as ExAC (see ExAC) and contained exome data only.
For more information, visit http://gnomad.broadinstitute.org.
1000 Genomes¶
The 1000 Genomes table provides information from the following populations:
AFR - Total African Ancestry population
AMR - Total Americas Ancestry population
EAS - Total East Asian Ancestry population
EUR - Total European Ancestry population
SAS - South Asian Ancestry population
TOTAL - Total variant allele frequency in the combined populations
Information provided by the 1000 Genomes Project about population survey sizes is shown in the following table:
Population |
Total |
|---|---|
Total African Ancestry (AFR) |
691 |
Total Americas Ancestry (AMR) |
355 |
Total East Asian Ancestry (EAS) |
523 |
Total European Ancestry (EUR) |
514 |
Total South Asian Ancestry (SAS) |
494 |
TOTAL |
2,577 |
For more information, visit 1000 Genomes Project phase 3.
EVS¶
The EVS table provides information from the following populations, derived from the NHLBI Exome Sequencing Project (ESP):
AFAM - African American population
EUAM - European American population
For more information, visit http://evs.gs.washington.edu/EVS/
Genome of Iceland¶
The Genome of Iceland table provides the following information from the Icelandic population:
TOTAL - Total allele frequency in the population
The Icelandic population dataset contains whole genome sequences from 2.636 individuals from Iceland. This project was executed by deCODE genetics (Gudbjartsson et al, 2015).
For more information, visit https://www.ncbi.nlm.nih.gov/pubmed/25807286.
Genome of the Netherlands¶
The Genome of the Netherlands table provides the following information from the Dutch population:
TOTAL - Total allele frequency in the population
The Genome of the Netherlands (GoNL) dataset includes 769 samples (The Genome of the Netherlands Consortium, 2014).
For more information, visit http://www.nlgenome.nl.
Human Genetic Variation Database (Kyoto - Japan)¶
The Human Genetic Variation Database (HGVD) table provides the following information from the Japanese population:
TOTAL - Total allele frequency in the population
The Kyoto Japanese population dataset includes 1,208 samples (Higasa et al, 2016).
For more information, visit http://www.hgvd.genome.med.kyoto-u.ac.jp/about.html.
Rotterdam Study Exome Sequencing¶
The Rotterdam Study Exome Sequencing table provides the following information from the Rotterdam Study, a prospective cohort study in Rotterdam, the Netherlands, ongoing since 1990:
TOTAL - Total allele frequency in the population
For more information, visit https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2071967/.