Managing Genelists¶
On the Advanced report settings tab, commonly used gene lists may be selected for analysis of a study. However, in some instances a custom gene list is desired. Gene lists can be used to collect and save lists of genes. They are associated with your user account and may be applied to any study within a project.
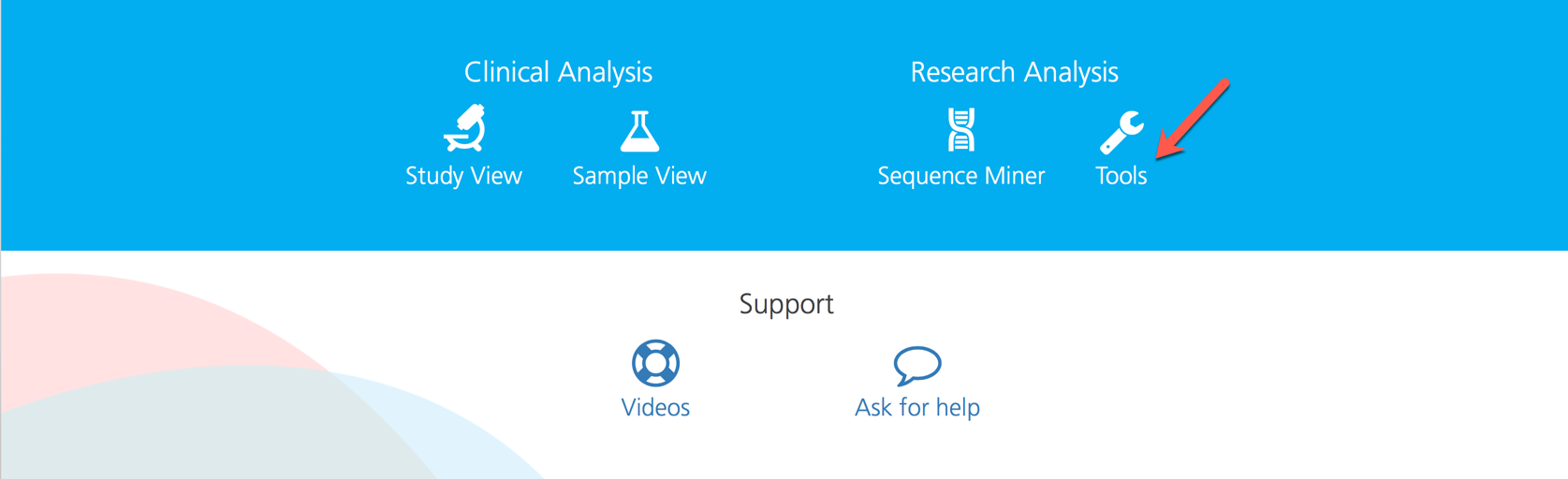
User-defined custom gene lists are managed from the User-defined genelists page, which is accessed by clicking the Tools link on the CSA dashboard.

Creating a genelist file¶
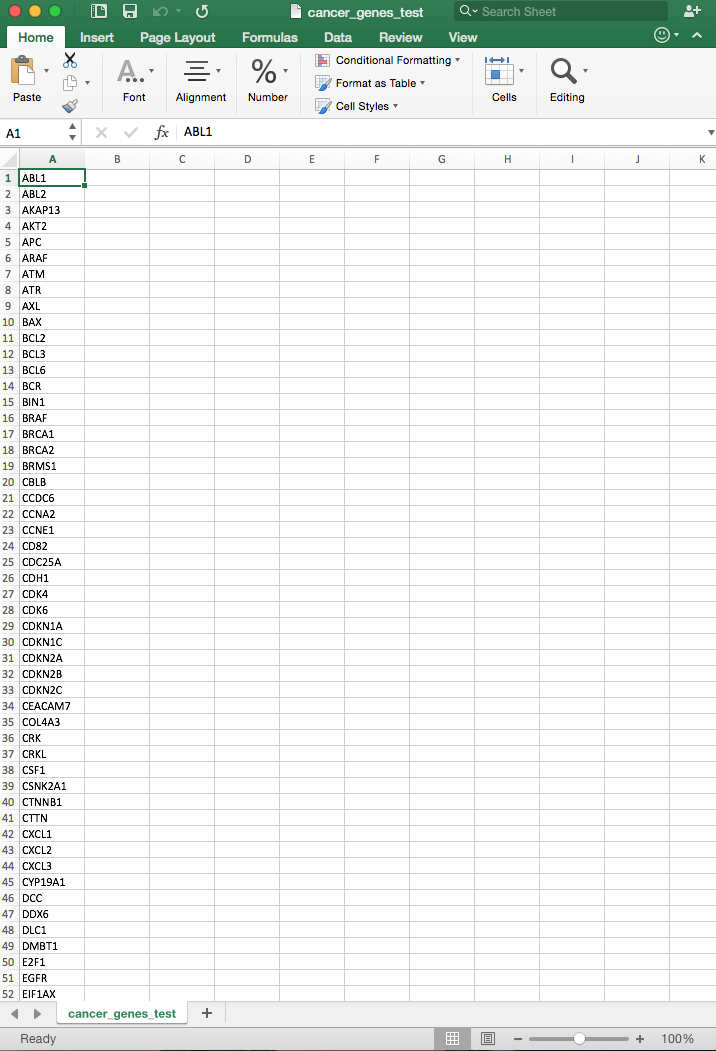
A user-defined genelist file consists of a tab-delimitted file (e.g., a text file) that contains one gene symbol per line. The file should NOT include a header row.
File contents¶
The gene symbols in a genelist are HUGO Gene Nomenclature Committee (HGNC)-approved gene symbols. These are the primary human gene names used by NCBI and Ensembl.
File format¶
You can create the file in a text editor, spreadsheet, or word-processing program. If you are using a spreadsheet to create the file (as shown below), save the file in a tab-delimited text format. If you are using a word-processing program, save the file as plain text. Once the file is saved to your computer, you can upload it to CSA.
Uploading a genelist¶
To upload a new genelist, click the Tools link on the CSA dashboard to open the User-defined genelists page.
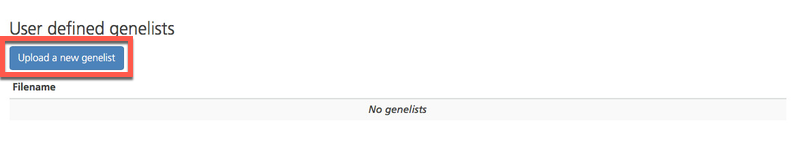
At the top of the page, click Upload a new genelist.
On the Create genelist page, click Choose file and select the file. Once the file is selected, the file name appears next to the Choose file button.
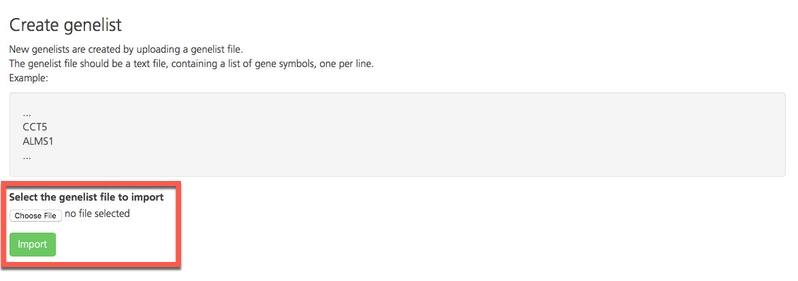
Click Import to upload and verify your file.
Verifing the genelist¶
Upon import, the gene symbols provided in the uploaded genelist file are verified by checking them against the HGNC database and displayed on the Verify genelist page. If a match is found, a checkmark appears next to the matched gene symbol in the Matched genes column. If a match is not found, “None” is displayed in the Matched genes column. If a gene appears more than once in the upload file, the number of replicates found in the file is displayed in parentheses next to the gene symbol.
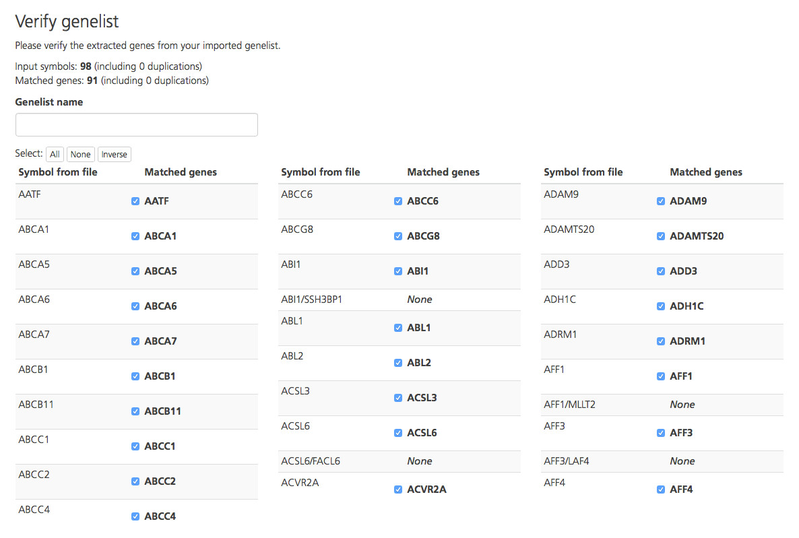
To verify that the gene symbols in the uploaded file correspond correctly to those in the Matched genes column, click the checkmark. You can select or omit all genes, a subset of genes, or none of the genes.
In the Genelist name field at the top of the page, enter a unique name for the new genelist.
Once all desired genes have been selected, click Verify and submit to complete the import. The file is saved in CSA as a .set file. If a name was not entered for the genelist, the genelist is automatically named “.set”.
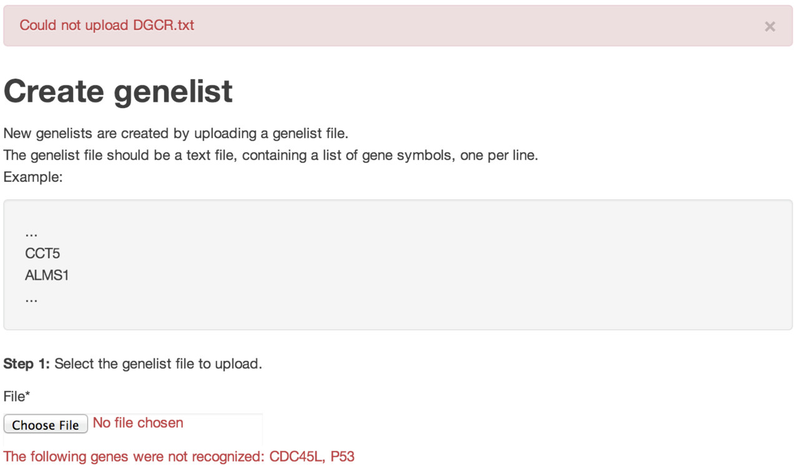
If the upload failed, a red banner displays at the top of Create genelist page to notify you that the file could not be uploaded. Failure occurs if one or more gene symbols are not recognized or if the file is not correctly formatted (e.g., if the file contains additional text fields). If failure occurs, correct the file and upload again.
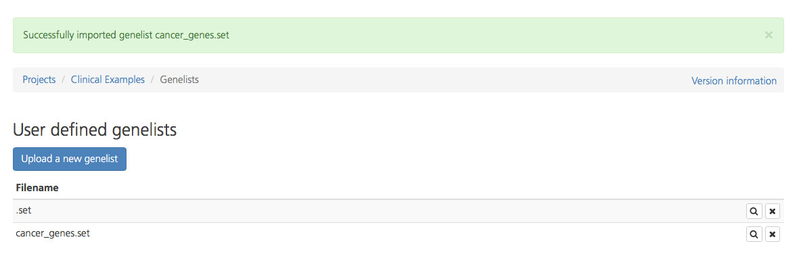
If the upload was successful, the new genelist appears on the User-defined genelist page and is available to view or to select when defining reportSettings.
Viewing a genelist¶
To view a user-defined custom genelist, click the Tools link on the CSA dashboard to open the User-defined genelists page.

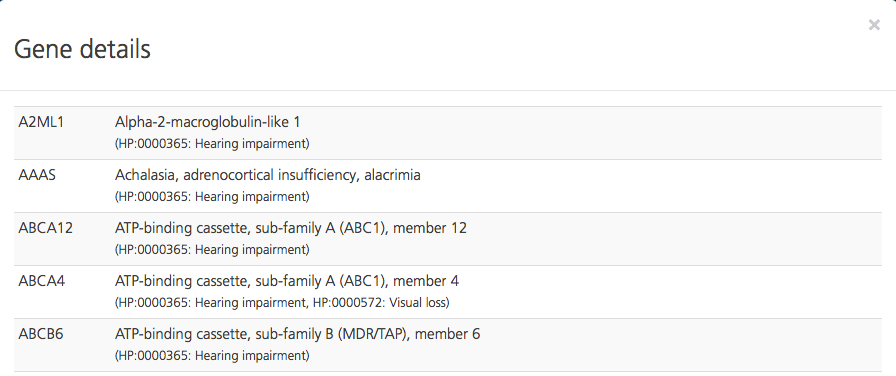
Genelists are listed in the order that they were uploaded, with the most recent at the top. To view a genelist, click the View gene details (magnifying glass) icon on the right-hand side of the selected row.

The Gene details pop-up window opens, which shows the gene symbols and gene names contained in the selected genelist.
Deleting a genelist¶
To delete a genelist, click the Delete (x) icon on the right-hand side of the selected row.

In the Remove genelist dialog, click Yes to confirm.

Note
Deleted genelists cannot be recovered.