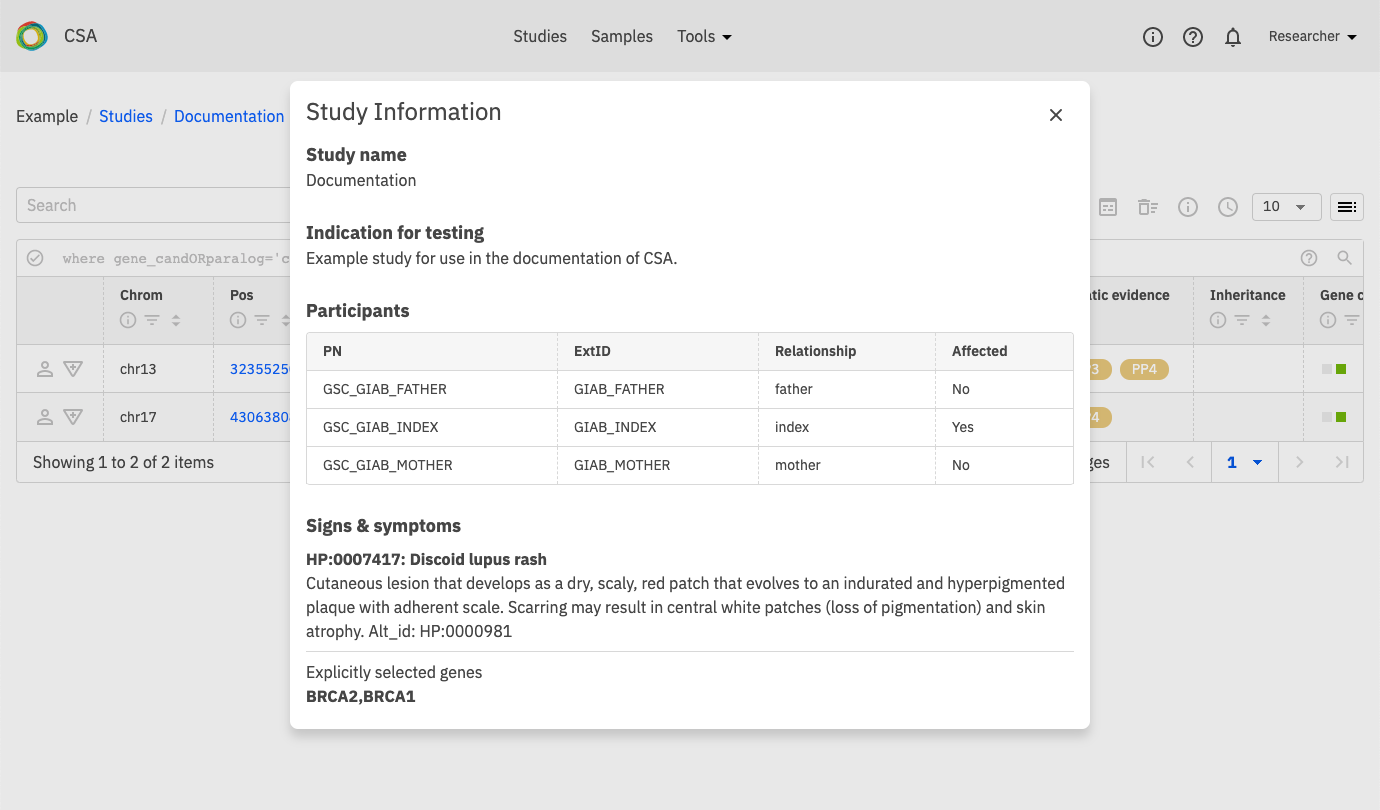
Variants¶
The Variants page displays all variants in a study based on the query settings for that study. There are three predefined filters shown on the variant page for Candidate Genes, OMIM and All Variants.
When the Variants page is first loaded, it takes some time to run the initial queries for the variant information for the study. This can take approximately ten minutes the first time you open the page. If you click on the trash icon on the Variants page, the query results will be invalidated and reloaded, which will require this initial loading time to be repeated.
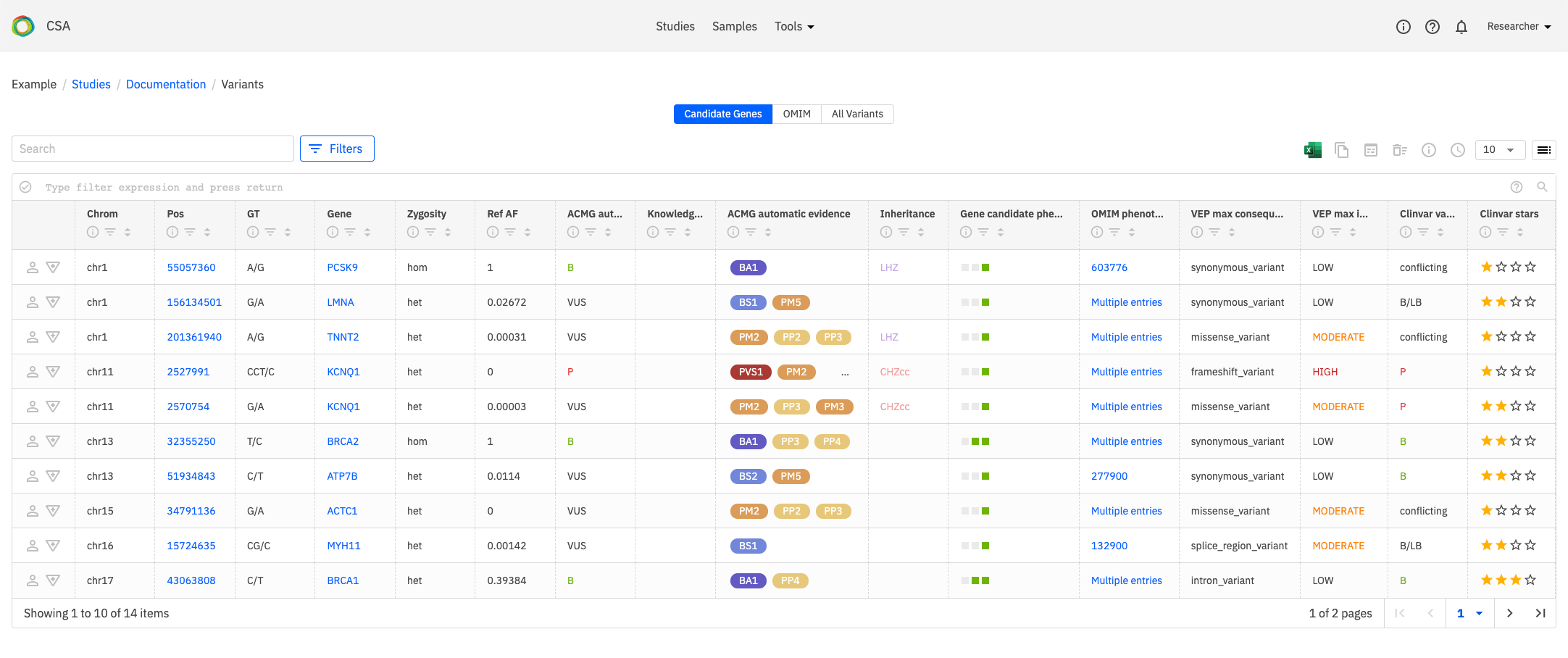
Candidate Genes¶
The default filter only shows variants that are associated with the candidate genes that have been associated with the study.
These candidate genes can be edited from the study’s query settings, which are shown in the image below. A list of candidate genes are shown in Selected candidate genes. Other genes can be added to the study by typing the name of the gene in the provided text box (indicated by the orange box below). A search box for phenotypes, signs or symptoms allows you to find genes to add to the candidates.
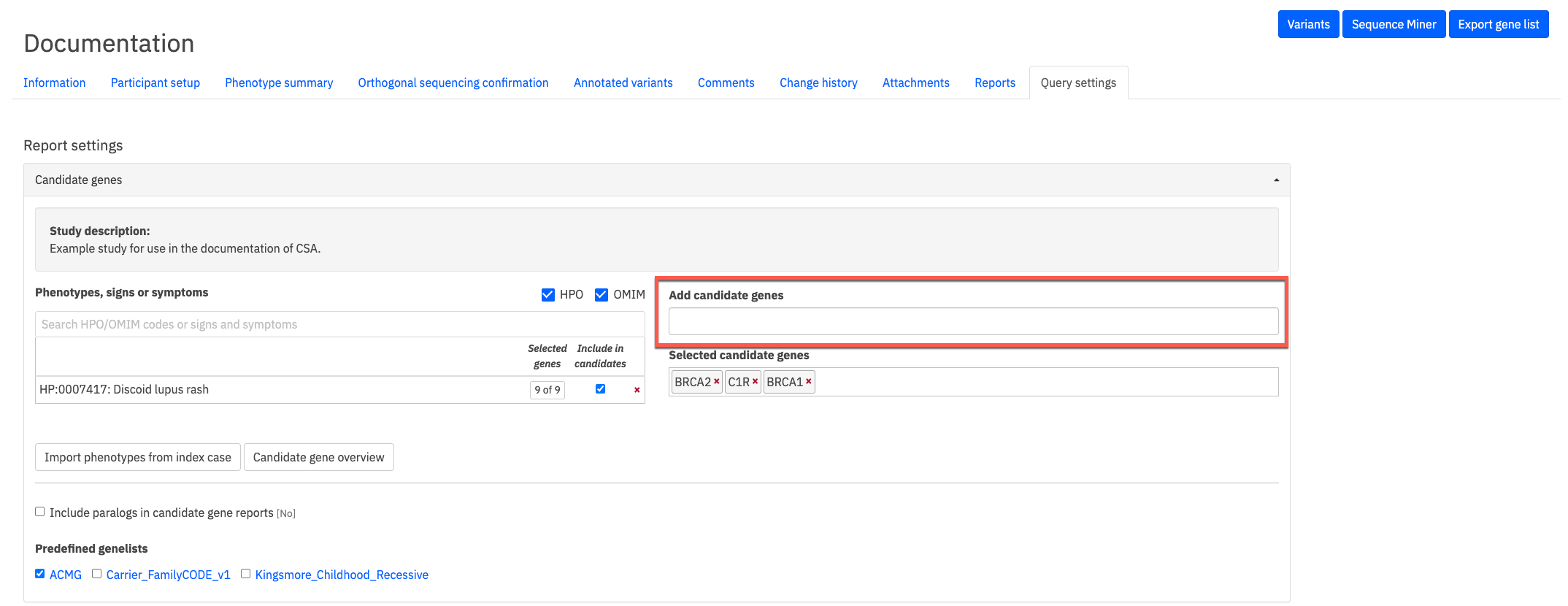
Selecting any of the options for ACMG, Carrier_FamilyCode_v1 or Kingsmore at the bottom of this view will add those genes to the candidates genelist.
Filters¶
A set of predefined filters can be selected by clicking the Filters button next to the Search field. The Filters dialog is shown below.
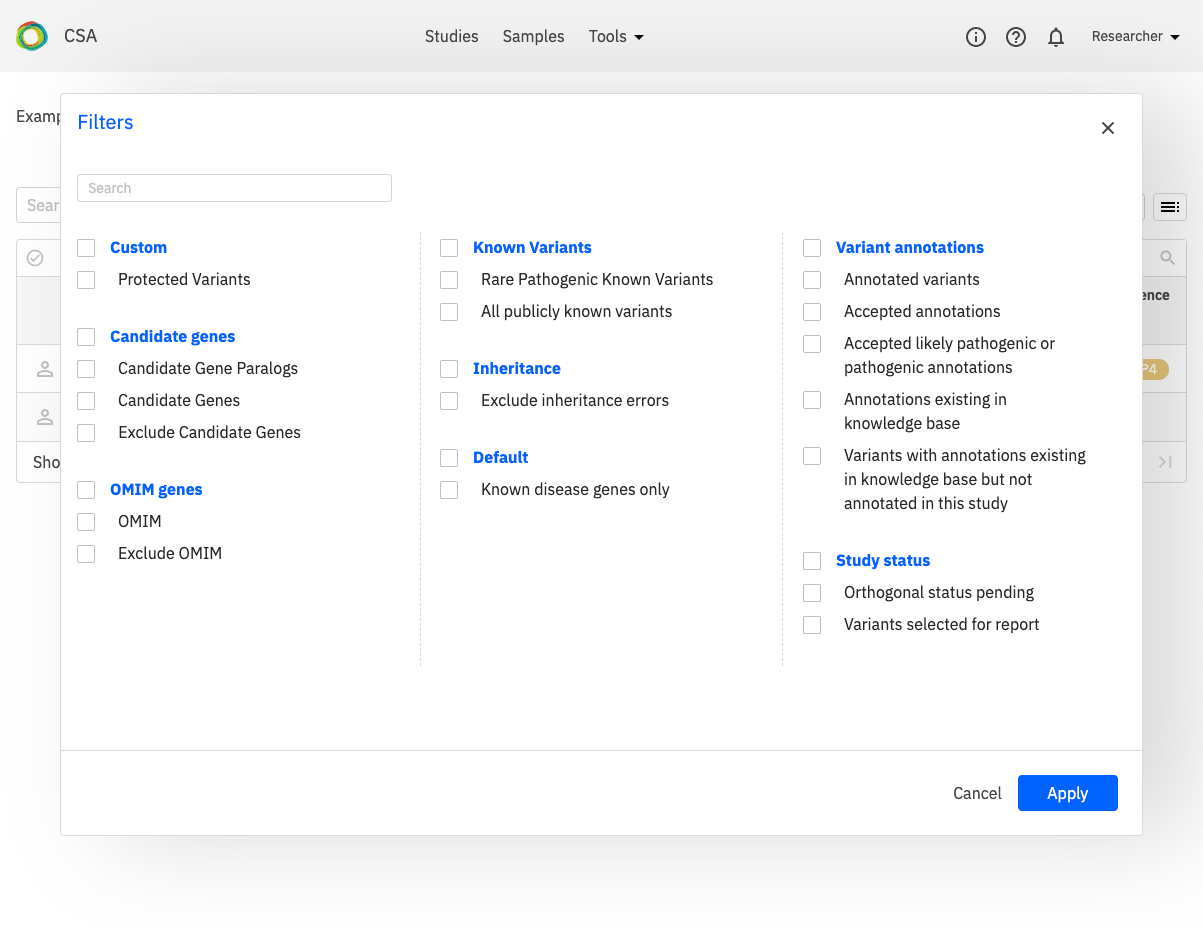
When a filter has been selected, it will be added to the filter expression input field.
Filters can also be set on each of the columns in the display.
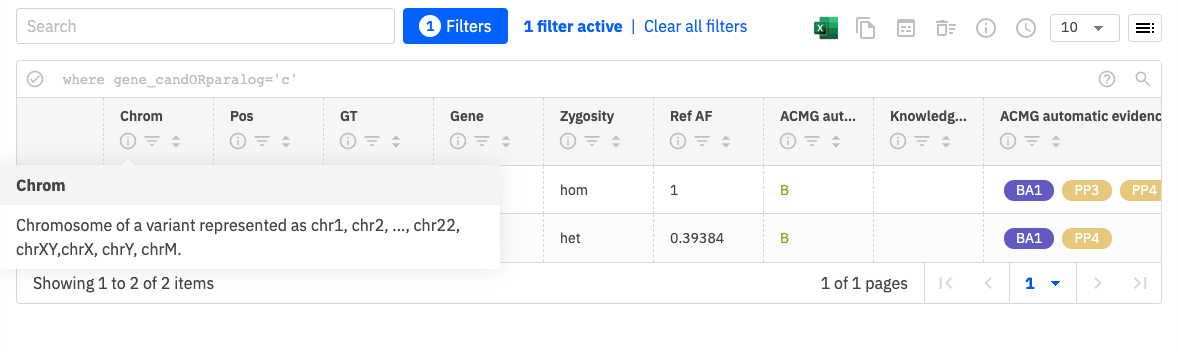
In addition to filtering and sorting options, you can click on the information icon for each column to display information about the content of that column, as shown here.
Variant Toolbar¶
You can export data and perform various other tasks on the Variants page using the toolbar on the Variants page, which is shown in the orange box below.
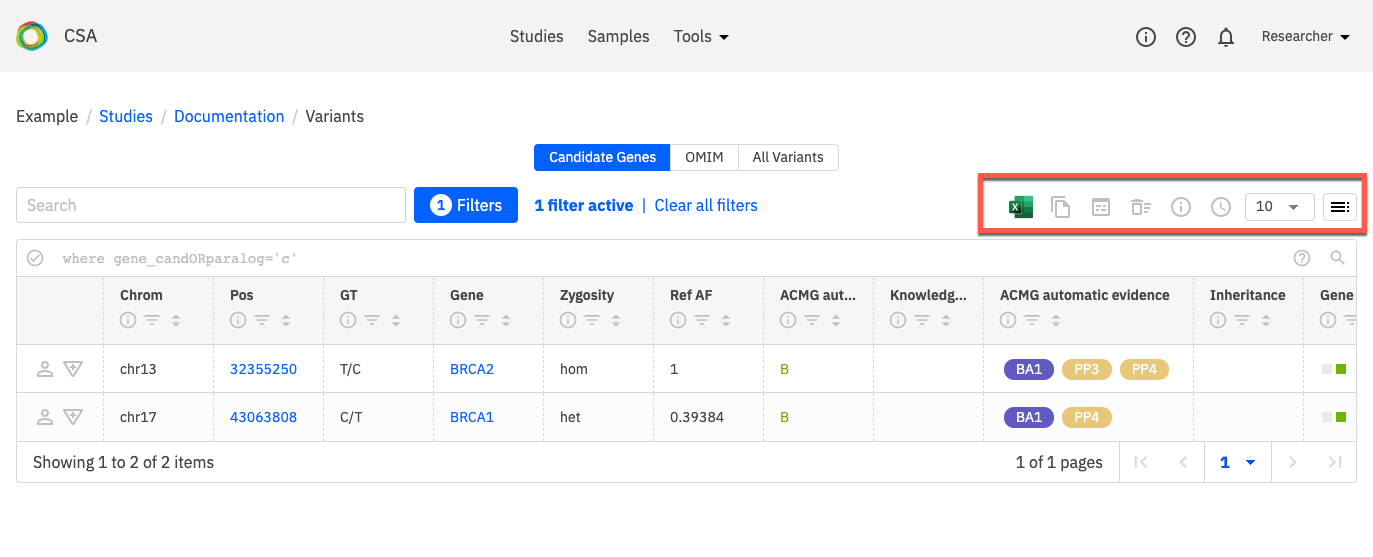
Export to Excel - You can export the currently viewed table to an Excel workbook. If you wish, you can export all columns (including those that are not currently in view) by selecting All columns from the menu provided.
Copy GOR query - The GOR query behind the table that is being viewed can be copied to the clipboard.
Open in Sequence Miner - The table can be opened in Sequence Miner (opens the genome browser to the chromosome and position that is in view).
Clear the cache - If you wish to clear the cached information behind the tables, the variants page will be re-initialised. Note that it will take another 10 to 15 minutes to run the GOR queries behind this table when the cache is cleared.
Show study information - Show various information about the study including: study name, indication for testing, signs & symptoms and explicitly selected genes in the study.
View query information - View information on when the query was run and how long it took to run.
Set number of rows to display
Column chooser - Select the columns that you wish to display in the view. Note that this does not persist beyond the current session and your preferences will be reset if you clear your browser data at any point.